Figure 1.
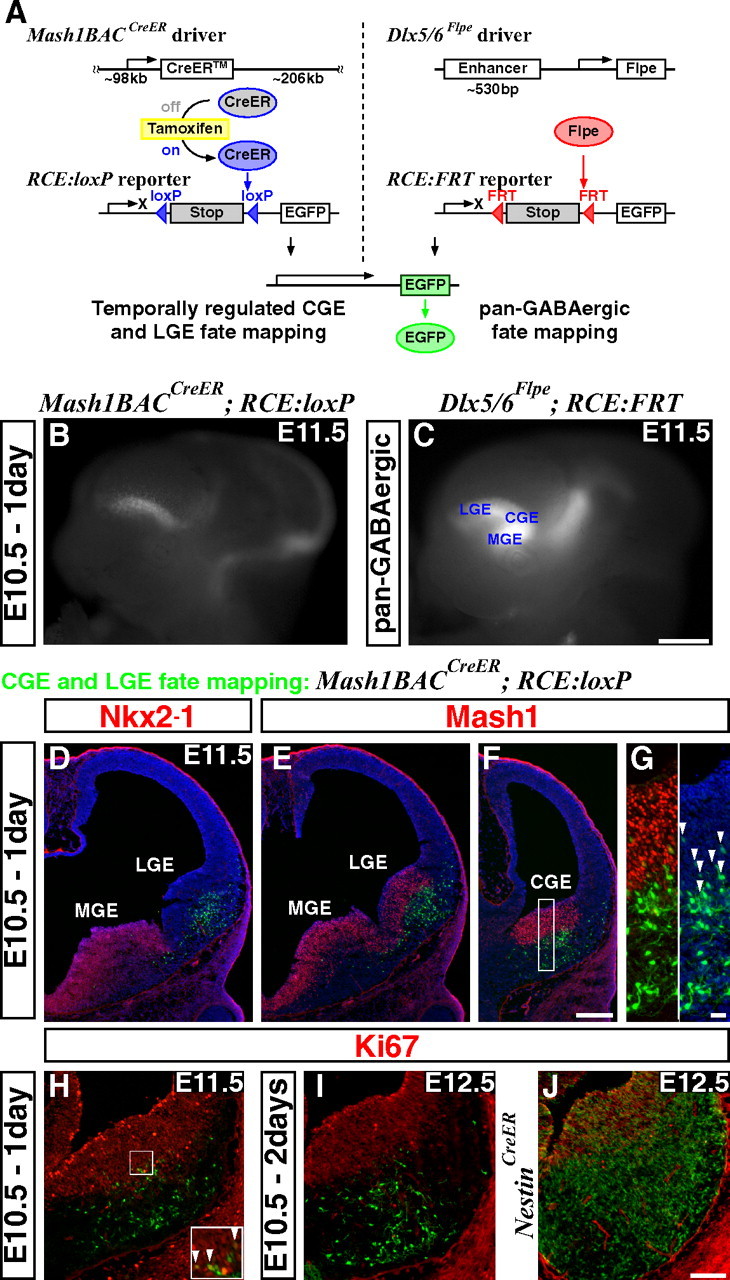
An inducible genetic strategy for fate mapping temporally distinct interneuron cohorts derived from the CGE. A, Schematics of an inducible genetic fate mapping strategy for CGE- and LGE-derived cells (left), and a method for fate mapping the entire GABAergic population in the forebrain (right). Left, By combining a Mash1BAC-CreER driver with an RCE:loxP reporter, Mash1-expressing cells in the CGE and LGE can be labeled at the desired time point by administration of tamoxifen. Within 6–24 h of tamoxifen administration, CreER is activated and removes the stop cassette in the RCE:loxP reporter by recombining the flanking loxP sites, resulting in permanent EGFP labeling of the cells expressing Mash1 in the CGE and LGE at this time. Right, Our Dlx5/6-Flpe driver expresses the site-specific recombinase Flpe under the regulation of the intergenic enhancer between Dlx5 and Dlx6 (id6/id5). By combining this line with the RCE:FRT reporter, which possesses a stop cassette flanked by FRT sites, the entire forebrain GABAergic population is labeled with EGFP. Note that both RCE reporters (RCE:loxP and RCE:FRT) were generated from a single dual-stop reporter (RCE:dual; see supplemental Fig. 5, available at www.jneurosci.org as supplemental material). B, EGFP expression in E11.5 whole mount brains after E10.5 tamoxifen administration. Note that EGFP expression is excluded from the MGE (compare with C). In addition to the labelings in the LGE and CGE, EGFP expression was observed in the thalamus and mid/hindbrain regions. C, Dlx5/6-Flpe directed pan-GABAergic EGFP labeling in the forebrain region is observed throughout the MGE, CGE and LGE, as well as in the thalamus at E11.5. D–G, Coronal telencephalic sections of B. D, As suggested by whole mount analysis (B), EGFP labeling is excluded from the MGE, which is marked by Nkx2–1 expression. E, F, Although Mash1 is normally expressed throughout the entire ventral forebrain progenitor zone (E, F), this particular founder does not recombine the reporter in the MGE. This allowed us to specifically label the cells in the CGE (F) and the LGE (E). G, Higher magnification of the CGE progenitor domain of F. All of the EGFP-positive cells coexpressing Mash1 (arrowheads) show only low levels of EGFP expression, suggesting that the removal of the stop cassette in the reporter has occurred only recently in these cells. H, A section adjacent to F. Very few proliferating cells (Ki67-positive) are labeled 1 d after tamoxifen administration (arrowheads in inset). I, Two days after tamoxifen administration, no fate-mapped cells coexpress Ki67. J, By comparison, many of the cells fate mapped using a Nestin-CreER driver line continue to proliferate 2 d after tamoxifen administration. Scale bars: B, C, 500 μm; D–F, 200 μm; G, 20 μm; H–J, 100 μm.
