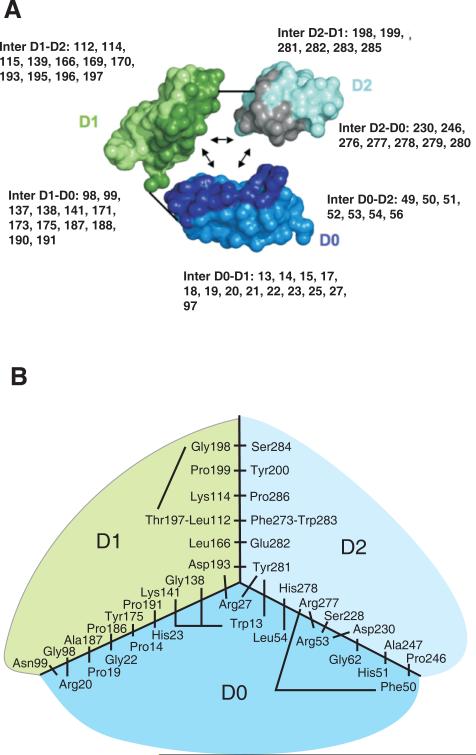
Figure 7. Model for the interaction of the D0, D1 and D2 domains in KIR3DL1.
Panel A shows the interactions between D0, D1 and D2 in the structural model. KIR3DL1 is represented in a simplified way with each amino acid being represented by two balls, one corresponding to the Cα atom the other to the center of the side-chain. The three domains have been separated so as to show their mutually interacting surfaces, which are shaded more darkly. The short straight lines show where the polypeptide chain connects D0 to D1 and D1 to D2. In each domain, the residues that contribute to interdomain interactions are shown. Here panel B shows a schematic view of the interacting residues that bring the three domains together. The view is from the top of the ligand-binding site in a two dimensional projection. The central part of the diagram corresponds to the junction of the three domains D0, D1 and D2. Connecting lines represent interactions between pairs of residues. Not all hydrophobic residues from the hydrophobic core are shown; those absent are valine 145, alanine 169, glycine 170, valine 196.