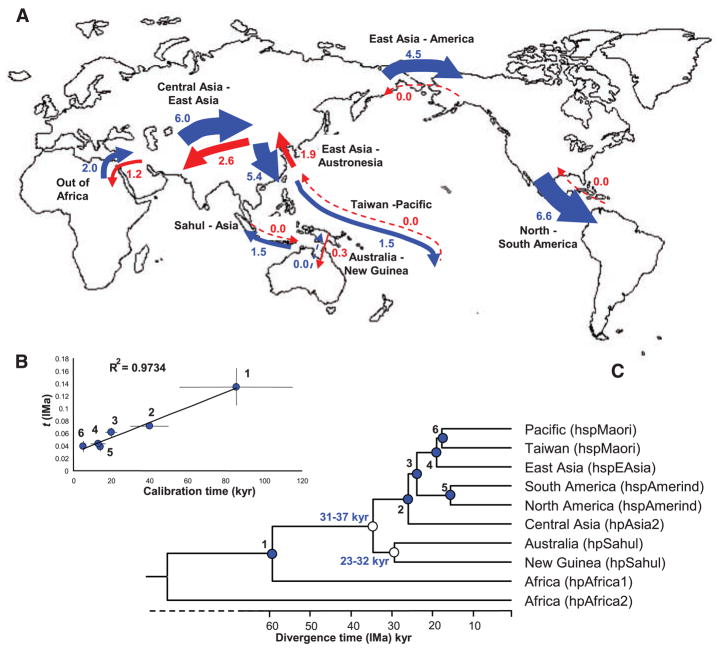
Fig. 2.
Global patterns of migration between eight pairs of H. pylori populations as calculated by the isolation with migration model (IMa). (A) Map. The magnitudes of migration are denoted by numbers and arrow thickness and their direction is indicated in blue or red. (B) Graph showing a linear relation between the calibration time (table S2) of six events (filled blue circles) that are dated by archaeological estimates and the estimated time (t). (C) Population tree reconstructed from a consensus of 1000 bootstrap samples from the range of calculated t values to determine the ages of nodes (thousands of years, kyr) associated with the peopling of the Sahul (unfilled circles). Ages (in light blue) are the 95% confidence limits of estimated coalescence times obtained by applying global rate minimum deformation (GRMD) rate-smoothing, as implemented in Treefinder, to the range of t values within the limits of calibration dates.