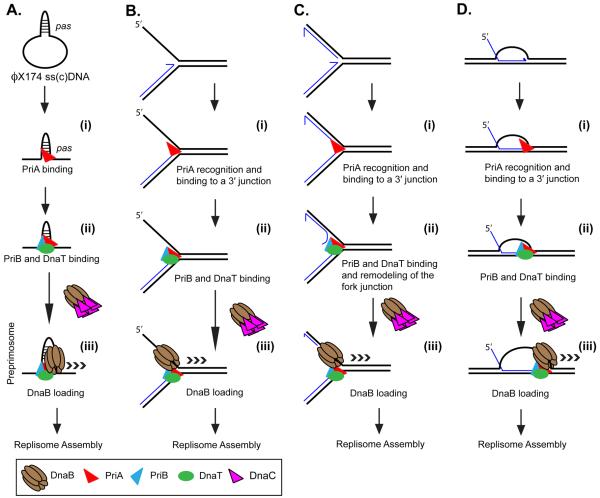
Figure 1.
PriA replisome loading. PriA can load a replisome to various DNA structures. (A) A primosome assembly site on ϕX174 viral DNA. (B) A stalled replication fork where there is a gap in the nascent lagging-strand. Such a structure can form when a leading-strand template block terminates all DNA synthesis. (C) A stalled replication fork where both template strands have been completely copied up to the point of template damage. Similar to (B) above, such a structure may occur when the replisome encounters a leading-strand blockage. Replisome loading on such a structure requires the unwinding of a portion of the nascent lagging-strand DNA to generate a single-stranded region for DnaB binding. (D) A recombinant joint molecule, represented here as a D loop. These structures necessarily occur for restart after the encounter of a template double-strand break and may also be generated under other situations as well. Although the DNA structures are different, the steps in loading the replisome are identical. (i) PriA recognizes the 3′ OH end of the nascent leading-strand DNA at the three-strand junction and (ii) recruits other accessory factors that may be important in fork remodeling (for example, in panel C). Recognition of the pas probably occurs through interaction of PriA with the psuedo-three stand junction formed by the hairpin in the DNA and the adjacent single-stranded DNA. (iii) An interaction between DnaT and DnaC, the helicase loader, allows loading of the replicative helicase, DnaB, forming the preprimosome (composed of PriA, PriB, DnaT, and DnaB). Once loaded, DnaB recruits DnaG via a protein-protein interaction. Synthesis of a primer then attracts the DNA polymerase III holoenzyme and a protein-protein interaction between the τ subunit of the holoenzyme and DnaB cements formation of the replisome. Nascent template 3′ OH ends are denoted by half-arrows.