Abstract
Phenylketonuria is an autosomal recessive disorder caused by a deficiency of phenylalanine hydroxylase. Transthyretin has been implicated as an indicator of nutritional status in phenylketonuria patients. In this study, we report that phenylalanine and its metabolite, phenylpyruvic acid, affect MAPK, changing transthyretin expression in a cell- and tissue-specific manner. Treatment of HepG2 cells with phenylalanine or phenylpyruvic acid decreased transcription of the TTR gene and decreased the transcriptional activity of the TTR promoter site, which was partly mediated through HNF4α. Decreased levels of p38 MAPK were detected in the liver of phenylketonuria-affected mice compared with wild-type mice. In contrast, treatment with phenylalanine increased transthyretin expression and induced ERK1/2 activation in PC-12 cells; ERK1/2 activation was also elevated in the brainstem of phenylketonuria-affected mice. These findings may explain between-tissue differences in gene expression, including Ttr gene expression, in the phenylketonuria mouse model.
Keywords: hepatocyte nuclear factor 4, mitogen-activated protein kinases, mouse, phenylketonurias, prealbumin
Introduction
Phenylketonuria (MIM 261600) is an autosomal recessive metabolic disorder in which insufficient phenylalanine hydroxylase (PAH; EC 1.14.16.1) activity results in the accumulation of phenylalanine and its metabolites in the blood and other organs. High levels of phenylalanine and its metabolites cause cognitive dysfunction, low brain weight, and changes in myelin structure (Scriver et al., 2001; Martynyuk et al., 2005). As phenylketonuria patients should consume a phenylalanine-restricted diet from birth to prevent brain dysfunction, it is important to monitor their nutritional status (Scriver et al., 2001). The serum level of transthyretin (MIM 176300), formerly called prealbumin, has been implicated as an indicator of the nutritional status of phenylketonuria patients (Shenton et al., 1983; Acosta et al., 1999; Arnold et al., 2002).
Transthyretin is a 55-kDa protein consisting of four identical subunits of 127 amino acids each (Episkopou et al., 1993). Transthyretin is the primary carrier of thyroid hormones and retinol and is mainly synthesized in the liver and choroid plexus (Episkopou et al., 1993). Transthyretin has been associated with amyloidosis, including familial amyloidotic polyneuropathy and Alzheimer's dementia (Costa et al., 1978; Choi et al., 2007; Buxbaum et al., 2008). In addition, transthyretin is involved with many acute-phase proteins when the liver initiates an acute-phase response to acute injury or infection (Wang and Burker, 2007). As serum transthyretin level has been implicated as an indicator of nutritional status in phenylketonuria patients, a better understanding of the regulation of TTR gene expression by phenylalanine and its metabolites is essential (Shenton et al., 1983; Arnold et al., 2002).
The Ttr gene is transcriptionally regulated by two DNA regions: a proximal -150 to -90 bp promoter region and a distal 100-nucleotide enhancer located -2 kb upstream of the mRNA cap site (Samadani and Costa, 1996). The Ttr promoter and enhancer regions have been identified in experiments with transgenic mice, and have DNA binding sites for four hepatocyte nuclear factors (HNFs), CCAAT/ enhancer binding protein (C/EBP), and AP-1/cJun (Costa and Grayson, 1991). More specifically, the Ttr proximal promoter region has binding sites for HNF1, HNF3, HNF4, HNF6, and AP-1. The Ttr enhancer region has binding sites for HNF3, HNF4, and C/EBP.
The mouse model of phenylketonuria (BTBR-Pahenu2) has a homozygous F263S mutation of the Pah gene, which is created by an alkylating agent, N-ethyl-N- nitrosourea (McDonald et al., 1990; McDonald and Charlton, 1997). The phenotype of the phenylketonuria mouse model closely resembles that of phenylketonuria patients: hypopigmentation, elevated serum and brain phenylalanine levels, and neurological abnormalities (McDonald et al., 1990; Surendran et al., 2004). Previously, we found that Ttr gene expression was downregulated in the liver and serum of phenylketonuria-affected mice compared with wild-type mice, although all mice were given ad libitum access to a diet containing phenylalanine (Park et al., 2009). In this report, we describe the mechanism by which phenylalanine and its metabolite, phenylpyruvic acid, regulate transcription of the Ttr gene in phenylketonuria.
Results
Phenylalanine and phenylpyruvic acid treatment of HepG2 cells reduces TTR mRNA and inhibits the activity of the TTR gene promoter
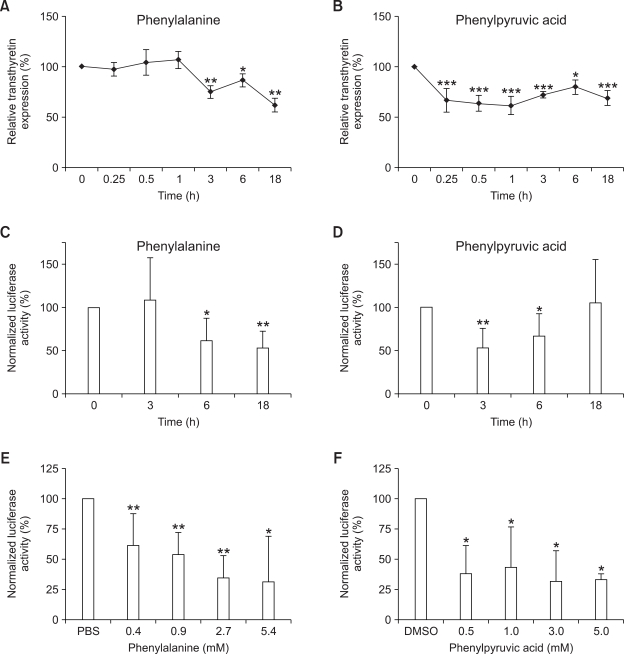
We performed real-time PCR analysis to determine whether the TTR mRNA level is altered by phenylalanine or phenylpyruvic acid treatment. Exposure of HepG2 cells to 0.9 mM phenylalanine reduced TTR transcription by 24.8 ± 6.5% after 3 h (P < 0.01), by 13.5 ± 6.5% after 6 h (P < 0.05), and by 38.3 ± 6.6% after 18 h compared with the control (P < 0.01; Figure 1A). Treatment of HepG2 cells with 1.2 mM phenylpyruvic acid resulted in a similar suppression of TTR transcription, except for a shift in the time course: transcription decreased to 66.6 ± 11.9% of that of the control after 15 min, and maximal repression of transcription was observed after 1 h (P < 0.001; Figure 1B).
Figure 1.
Real-time PCR analysis showing decreased TTR gene transcription in HepG2 cells treated with 0.9 mM phenylalanine (A) or 1.2 mM phenylpyruvic acid (B). HepG2 cells were transfected with a reporter plasmid containing the mouse Ttr promoter region (C-F). Twenty-four hours after transfection, 0.9 mM phenylalanine (C) or 1.2 mM phenylpyruvic acid (D) were added for the indicated times. To determine whether the effects of phenylalanine and phenylpyruvic acid were dose dependent, various concentrations of phenylalanine (E) or phenylpyruvic acid (F) were added. The cells were lysed, and firefly and Renilla luciferase activities were measured. Reporter activity was calculated as the ratio of firefly luciferase activity to Renilla luciferase activity in the same sample. Values are expressed as a percentage of the reporter activity relative to that of control cells, and are plotted as the mean ± SD (n = 4-6). *P < 0.05, **P < 0.01, ***P < 0.001 vs control cells using Student's t-test.
To assess the effects of phenylalanine and phenylpyruvic acid on Ttr promoter-mediated transactivation potential, a luciferase assay using a luciferase reporter containing a mouse Ttr promoter site was performed. As phenylalanine and phenylpyruvic acid dissolve PBS and DMSO, respectively, PBS- and DMSO-treated cells were used as controls. Exposure of HepG2 cells to 0.9 mM phenylalanine decreased luciferase activity by 38.2 ± 24.9% after 6 h (P < 0.05) and by 46.2 ± 18.9% after 18 h compared with the control (P < 0.01; Figure 1C). Luciferase activity decreased after 3 h and 6 h of 1.2 mM phenylpyruvic acid treatment (Figure 1D). Maximal repression occurred after 3 h, when luciferase activity was 53.1 ± 22.5% of that of the control (P < 0.01; Figure 1D). Increments in the concentration of phenylalanine induced marked decreases in luciferase activity in a dose-dependent manner (Figure 1E). In contrast, a small amount of phenylpyruvic acid (0.5 mM) resulted in a decrease in luciferase activity similar to that obtained with a greater amount of phenylpyruvic acid (P < 0.05; Figure 1F). These findings suggest that modification of TTR gene transcription by phenylalanine and phenylpyruvic acid is mediated though the TTR promoter region.
Phenylalanine and phenylpyruvic acid decrease the level of HNF4α protein in HepG2 cells, and the level of p38 MAPK is low in the liver of phenylketonuria-affected mice
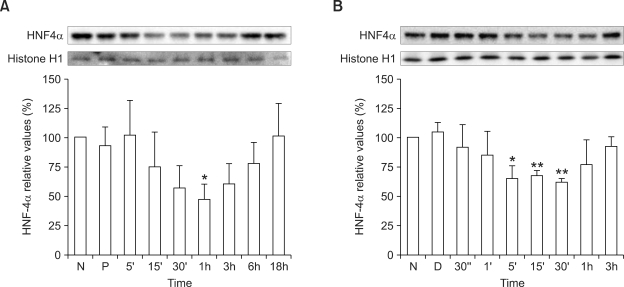
To identify transcription factors involved in phenylalanine- and phenylpyruvic acid-mediated TTR gene alteration, we analyzed protein levels of transcription factors that bind to the TTR promoter region. HNF4α nuclear protein level in HepG2 cells transiently decreased to 47.3 ± 13.0% of that of the control after 1 h of 0.9 mM phenylalanine treatment (P < 0.05; Figure 2A). The HNF4α nuclear protein level in HepG2 cells started to decrease after 5 min of 1.2 mM phenylpyruvic acid treatment and was 61.6 ± 2.8% of that of the control after 30 min (P < 0.01; Figure 2B). Treatment of HepG2 cells with 0.9 mM phenylalanine or 1.2 mM phenylpyruvic acid did not alter nuclear protein levels of HNF1, HNF3α/β, C/EBPα, or C/EBPβ(data not shown).
Figure 2.
HNF4α nuclear protein levels in HepG2 cells treated with 0.9 mM phenylalanine (A) or 1.2 mM phenylpyruvic acid (B). Values are expressed as percentages of values for untreated cells (N) and are plotted as the mean ± SD (n = 3). *P < 0.05, **P < 0.01 vs (A) PBS-treated control cells (P) or vs (B) DMSO-treated control cells (D) using Student's t-test.
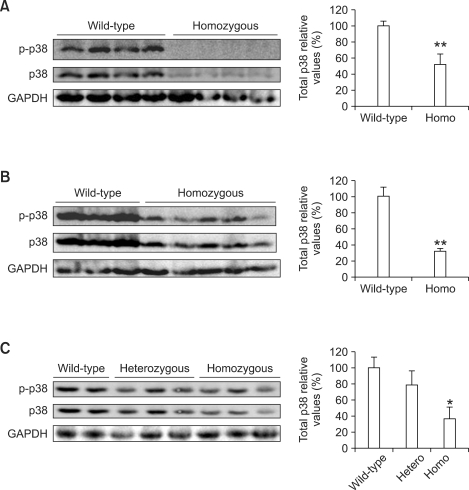
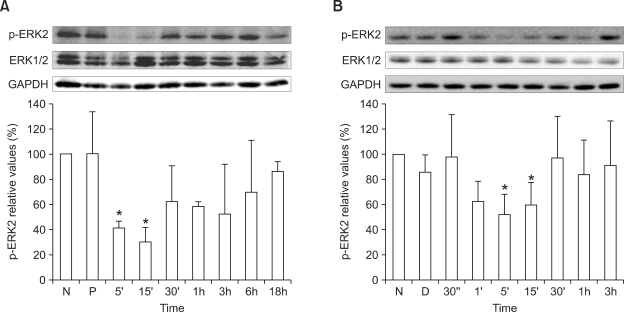
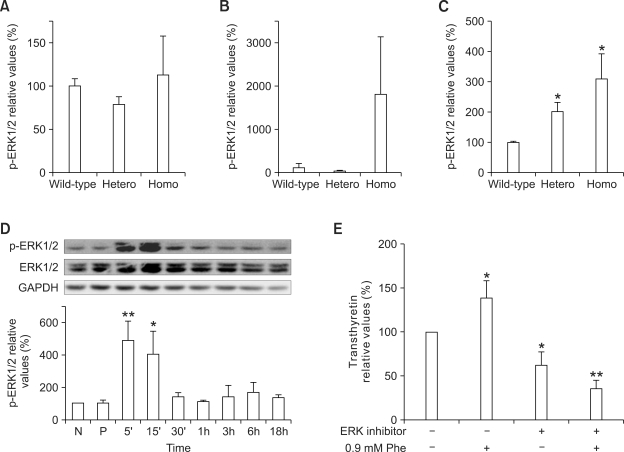
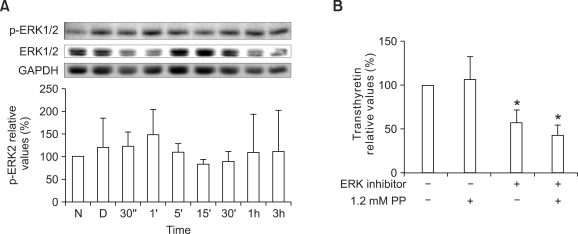
As MAPK has been reported to play an important role in the regulation of HNF4α activity (Hatzis et al., 2006; Li et al., 2006; Xu et al., 2007), we analyzed MAPK alterations in the liver of the phenylketonuria mouse model. Total levels of p38 MAPK protein in liver tissue were significantly lower in homozygous Pahenu2 mice than in wild-type mice: those of 5- week-old male homozygous mice were 52.2 ± 12.5% lower (P < 0.01; Figure 3A); those of 30- week-old male homozygous mice were 32.1 ± 3.2% lower (P < 0.01; Figure 3B); and those of 43-week-old female homozygous mice were 36.3 ± 14.9% lower (P < 0.05; Figure 3C). JNK/SAPK and ERK1/2 phosphorylation was not detected in liver tissues of the phenylketonuria mouse model (data not shown). We also monitored p38 MAPK and ERK1/2 activation in HepG2 cells treated with 0.9 mM phenylalanine or 1.2 mM phenylpyruvic acid. However, the phosphorylated form of p38 MAPK was not detected in HepG2 cells (data not shown). ERK2 inactivation occurred transiently in HepG2 cells after the addition of phenylalanine or phenylpyruvic acid (Figure 4).
Figure 3.
Western blotting was performed to detect p38 kinase expression in the liver of the phenylketonuria mouse model (BTBR-Pahenu2): (A) 5-week-old male mice, (B) 30-week- old male mice, and (C) 43-week-old female mice. Levels of p38 kinase protein in liver were quantified using a bioimaging analyzer. Values are expressed as the mean ± SD (n = 2-5). *P < 0.05, **P < 0.01 vs wild-type mice using Student's t-test.
Figure 4.
Western blotting analysis of HepG2 cells treated with 0.9 mM phenylalanine or with 1.2 mM phenylpyruvic acid. Whole cell lysates of HepG2 cells treated with 0.9 mM phenylalalnine (A) or with 1.2 mM phenylpyruvic acid (B) were used for detection of phosphorylated Erk2 and total Erk1/2. Values are expressed as percentages of values for untreated cells (N) and are plotted as mean ± SD (n = 3). *P < 0.05 vs (A) PBS treated control cells (P) or vs (B) DMSO treated control cells (D) using Student's t-test.
ERK1/2 is activated in the brainstem of phenylketonuria-affected mice
As brainstem transthyretin expression was previously shown to be elevated in homozygous mice, we examined MAPK alterations in brain tissues of the phenylketonuria mouse model (Park et al., 2009). ERK1/2 phosphorylation was elevated in the brainstem of heterozygous and homozygous mice compared with wild-type mice: by 100.0 ± 30.1% in heterozygous mice and by 207.2 ± 83.5% in homozygous mice (P < 0.05; Figure 5C). Although the level of the phosphorylated form of ERK1/2 increased greatly in the cerebellum of homozygous mice, the increase was not statistically significant (Figure 5B). ERK1/2 phosphorylation levels in the cerebrum were not statistically significant between groups (Figure 5A). JNK/SAPK and p38 kinase phosphorylation levels in the brain were not statistically significant between groups (data not shown).
Figure 5.
Western blotting was performed to detect ERK1/2 expression levels in the brain of the phenylketonuria mouse model (BTBR-Pahenu2). ERK1/2 protein expression in (A) the cerebrum, (B) the cerebellum, and (C) the brain stem was demonstrated and quantified using a bioimaging analyzer. Whole cell lysates of PC-12 cells treated with (D) 0.9 mM phenylalanine were used for detection of phosphorylated ERK1/2 and total ERK1/2. Western blotting was performed to detect transthyretin expression levels in PC-12 cells treated with (E) 0.9 mM phenylalanine after preincubation with ERK inhibitor II. The cells were exposed to phenylalanine for 7 days. Transthyretin levels were normalized to GAPDH levels. Values are expressed as the mean ± SD (n = 3-4). *P < 0.05, **P < 0.01 vs wild-type mice or DMSO- and/or PBS-treated controls using Student's t-test.
Phenylalanine-induced elevation of transthyretin expression in PC-12 cells is ERK1/2 dependent
We investigated whether phenylalanine and its metabolite, phenylpyruvic acid, affect ERK1/2 phosphorylation in PC-12 cells. Phosphorylated ERK1/2 protein levels increased transiently 5 min after treatment with 0.9 mM phenylalanine: by 390.4 ± 118.2% after 5 min (P < 0.01) and by 307.0 ± 142.0% after 15 min (P < 0.05; Figure 5D). ERK1/2 phosphorylation in PC-12 cells was not altered by treatment with 1.2 mM phenylpyruvic acid (Figure 5E). Exposure of PC-12 cells to 0.9 mM phenylalanine increased transthyretin expression by 38.6 ± 19.1% after 7 days (P < 0.05; Figure 5E) (Park et al., 2009). To examine the effect of ERK1/2 phosphorylation on transthyretin expression, PC-12 cells were pretreated with an ERK1/2 inhibitor. After pretreatment with ERK1/2 inhibitor II, the addition of 0.9 mM phenylalanine did not increase transthyretin expression in PC-12 cells (Figure 5E). The selective ERK1/2 inhibitor II decreased transthyretin expression in PC-12 cells regardless of treatment with 0.9 mM phenylalanine or 1.2 mM phenylpyruvic acid (Figure 6A, B). Transthyretin expression did not increase after treatment with 1.2 mM phenylpyruvic acid (Figure 6B).
Figure 6.
Whole cell lysates of PC-12 cells treated with (A) 1.2 mM phenylpyruvic acid were used for detection of phosphorylated ERK1/2 and total ERK1/2. Western blot analysis was performed to detect transthyretin expression levels in PC-12 cells treated with (B) 1.2 mM phenylpyruvic acid after preincubation with ERK inhibitor II. The cells were exposed to phenylpyruvic acid for 7 days. Transthyretin levels were normalized to GAPDH levels. Values are expressed as the mean ± SD (n = 3-4). *P < 0.05 vs DMSO-treated controls using Student's t-test.
Phenylalanine and phenylpyruvic acid may affect TTR transcription independently
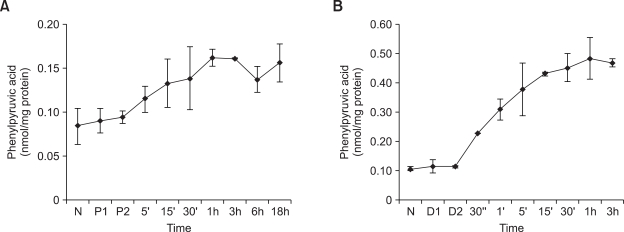
The effect of phenylpyruvic acid on suppression of TTR transcription in HepG2 cells was similar to that of phenylalanine, except for a shift in the time course. As phenylalanine may be transformed into phenylpyruvic acid, we hypothesized that phenylpyruvic acid, not phenylalanine, suppresses TTR transcription directly in HepG2 cells. We monitored levels of phenylpyruvic acid using GC/MS to determine whether phenylalanine was transformed into phenylpyruvic acid. Phenylpyruvic acid was not detected in phenylalanine-treated HepG2 cells (data not shown). As the PAH enzyme level was normal in HepG2 cells, the phenylalanine that was added may have been metabolized to tyrosine. Phenylpyruvic acid was detected in phenylalanine-treated PC-12 cells, which lack PAH enzyme activity (Figure 7A). However, the level of phenylpyruvic acid in phenylalanine-treated PC-12 cells was low compared with that in phenylpyruvic acid-treated PC-12 cells (Figures 7A and 7B). Phenylpyruvic acid in PC-12 cells reached a maximum concentration after 1 h of 0.9 mM phenylalanine treatment (Figure 7A), which is much longer than that required for phenylalanine to affect MAPK. These findings suggest that phenylalanine and phenylpyruvic acid affect TTR transcription independently.
Figure 7.
Phenylpyruvic acid levels in PC-12 cells treated with (A) 0.9 mM phenylalanine or (B) 1.2 mM phenylpyruvic acid assessed using GC/MS. Values are expressed as percentages of values for untreated cells (N) and are plotted as the mean ± SD (n = 2). P1 and P2 indicate PBS-treated control cells after 1 h and 6 h, respectively. D1 and D2 indicate DMSO-treated control cells after 1 min and 1 h, respectively.
Discussion
Previously, we detected elevated transthyretin expression in the brain and decreased transthyretin expression in the serum and liver of phenylketonuria-affected mice using a Pahenu2 phenylketonuria mouse model (Park et al., 2009). In addition, the transthyretin level decreased in HepG2 cells after treatment with 0.9 mM phenylalanine, a concentration typical of that in the plasma of phenylketonuria patients (Bedin et al., 2000; Park et al., 2009). The aim of this study was to investigate the mechanisms of differential transthyretin expression in the brain and liver of phenylketonuria-affected mice using cell lines. We used PC-12 cells as neuronal cells and HepG2 cells as liver cells. A deficiency of the hepatic enzyme, PAH, which converts phenylalanine to tyrosine, results in phenylketonuria. As phenylalanine cannot be oxidized to tyrosine in phenylketonuria, transamination of phenylalanine produces phenylpyruvic acid, which is further metabolized to phenyllactic acid and phenylacetic acid (Sarkissian et al., 2000). We used real-time PCR to examine the effect of phenylalanine and its metabolites on TTR transcription in HepG2 cells. Phenyllactic acid and phenylacetic acid did not alter TTR transcription significantly (data not shown). Only phenylalanine and phenylpyruvic acid induced a significant decrease in TTR transcription. The luciferase analysis with reporter plasmids inserted into the mouse Ttr promoter region revealed that modification of TTR gene transcription by phenylalanine and phenylpyruvic acid was mediated though the TTR promoter region. Of the transcription factors that bind to the TTR promoter region, the HNF4α nuclear protein level decreased. Reporter plasmids inserted into the mouse Ttr promoter region were used for luciferase analysis in HepG2 cells. The expression of a mouse gene in a human hepatoma cell line was successful because there is a high level of conservation between the mouse and human genes in the 190-nucleotide stretch just upstream from the cap site, and the general species barrier does not exist for liver-specific gene expression in HepG2 cells (Costa et al., 1989; Friedman et al., 1986).
A key role of HNF4α in the regulation of TTR gene transcription has been reported, and overexpression of HNF4α protein causes dose-dependent upregulation of TTR reporter gene transcription (Wang and Burker, 2007). In our study, nuclear HNF4α protein levels decreased in HepG2 cells treated with phenylalanine or phenylpyruvic acid. The phenylalanine- and phenylpyruvic acid-induced decreases in HNF4α level may have led to a decrease in TTR gene transcription in HepG2 cells. As we did not examine the effect of phenylalanine and its metabolites on the TTR enhancer region, this remains to be validated.
Several studies have suggested that there is a link between MAPK and HNF4α (Hatzis et al., 2006; Li et al., 2006; Xu et al., 2007). Inhibition of p38 kinase activity decreases HNF4α nuclear protein levels and its phosphorylation status in vivo and in vitro, which makes the protein less stable (Xu et al., 2007). Awad et al. (2000) and Mendelson et al. (1996) reported that p38 MAPK is constitutively active in the liver of adult mice, which is in accordance with our results. In our study, the p38 kinase protein level was significantly decreased in the liver of phenylketonuria-affected mice regardless of age or sex. As the total p38 kinase protein level and the level of the phosphorylated form of p38 kinase protein were greatly decreased in the liver of phenylketonuria-affected mice, this may have decreased HNF4α nuclear protein levels in phenylketonuria-affected mice. Suppression of HNF4α nuclear protein levels would diminish TTR gene transcription because HNF4α positively regulates TTR gene expression (Hatzis et al., 2006). ERK2 activation downregulates HNF4α expression in HepG2 cells at the transcriptional level via a mechanism that involves the loss of C/EBPa expression and the concomitant disruption of the HNF4 enhancer-promoter complex (Hatzis et al., 2006). ERK2 inactivation occurred transiently in HepG2 cells after the addition of phenylalanine or phenylpyruvic acid (Figure 4). As ERK2 activation decreases HNF4 expression, ERK2 inactivation may have increased HNF4α expression. However, neither phenylalanine treatment nor phenylpyruvic acid treatment altered C/EBPa nuclear protein levels in HepG2 cells (data not shown). In addition, HNF4α nuclear protein levels decreased after addition of phenylalanine or phenylpyruvic acid (Figure 2). Thus, altered ERK2 activation after addition of phenylalanine or phenylpyruvic acid may not have directly affected HNF4α expression in our study. Although we also performed electrophoretic mobility shift assays, we did not detect any alterations in the binding affinity of transcription factors, including HNF4α, in HepG2 cells treated with phenylalanine or phenylpyruvic acid (data not shown). Although phenylalanine and phenylpyruvic acid did not affect the binding affinity of HNF4α to the TTR promoter region, the decreased amount of HNF4α per se may have suppressed TTR gene transcription.
ERK1/2 activation was detected in the brainstem of 43-week-old homozygotes. In our previous study, increased transthyretin protein expression was detected in the brainstem of 30-week-old homozygotes (Park et al., 2009). To examine correlations between phenylketonuria, ERK1/2 activation, and transthyretin expression, PC-12 cells were treated with phenylalanine and phenylpyruvic acid. Although the PC-12 cell line does not represent brain tissue, it was used as one of the neuronal cell lines. Only phenylalanine, not phenylpyruvic acid, activated ERK1/2 in PC-12 cells. Exposure of PC-12 cells to phenylalanine increased transthyretin expression (Park et al., 2009). However, the addition of phenylalanine did not increase transthyretin expression in PC-12 cells after pretreatment with ERK1/2 inhibitor II. Thus, the elevated transthyretin expression in the brainstem of phenylketonuria-affected mice was probably the result of ERK1/2 activation induced by high brain phenylalanine levels. Of the transcription factors that bind to the TTR promoter region, only C/EBP and AP-1 exist in neuronal cells (Kuo et al., 1990). Thus, ERK1/2 activation by phenylalanine may have affected C/EBP and/or AP-1, which may have affected transthyretin expression in the brainstem of phenylketonuria-affected mice. The effect of phenylalanine and phenylpyruvic acid on C/EBP and AP-1 in the brain remains to be elucidated. Transthyretin has been associated with familial amyloidotic polyneuropathy (FAP), and ERK1/2 activation is upregulated in human FAP nerves and Schwann cells, whereas p38 and JNK MAPK activation is not detected in human FAP nerves and Schwann cells (Costa et al., 1978; Monteiro et al., 2006). Our data corroborate the results of Monteiro et al. (2006).
Serum transthyretin level has been implicated as an indicator of nutritional status in phenylketonuria patients (Shenton et al., 1983; Arnold et al., 2002). As phenylalanine and its metabolite, phenylpyruvic acid, decrease transthyretin expression in HepG2 cells, the use of serum transthyretin level as a nutritional indicator for phenylketonuria patients may be limited; further studies are necessary to elucidate this problem. In addition, further studies should be performed to validate the function of transthyretin in the phenylketonuria-affected brain.
In conclusion, our findings indicate that phenylalanine and its metabolite, phenylpyruvic acid, affect MAPK activation, resulting in tissue-specific transthyretin expression in a phenylketonuria mouse model. It has been suggested that the presence of cell-specific transcriptional factors would explain cell-specific regulation (Costa et al., 1989). However, the differential regulation of MAPK in specific cells or tissues remains to be elucidated. This may explain tissue-specific gene expression, including that of the TTR gene, and the tissue-specific vulnerability of the phenylketonuria mouse model.
Methods
Animals
The animal protocol was approved by the Animal Experiment Review Committee of the Ewha Medical Research Institute, Ewha Womans University, Korea. All animals were treated in accordance with the Animal Care Guidelines of the Ewha Womans University School of Medicine and the National Institutes of Health's Guidelines for Animal Care. The Pahenu2 phenylketonuria mice were obtained from the Jackson Laboratory (Bar Harbor, ME). The Pahenu2 phenylketonuria mouse contains a T-to-C transition in exon 7 of the PAH gene (McDonald et al., 1997). The genotype of each mouse was confirmed by genotyping. Mice were provided with water and given ad libitum access to a diet containing phenylalanine.
Cell culture
HepG2 and PC-12 cells were grown in DMEM (Gibco-BRL, Gaithersburg, MD) supplemented with 100 U/ml of penicillin, 100 µg/ml of streptomycin, and 10% (v/v) of heat-inactivated FBS (Gibco-BRL) at 37℃ in a humidified atmosphere containing 5% CO2. The medium was replaced with serum-free medium 6 h before the experiment. Each cell line was treated with 0.9 mM L-phenylalanine (Sigma- Aldrich, Madison, WI) or 1.2 mM phenylpyruvic acid (Fluka, Milwaukee, WI) in serum-free medium for the intervals indicated in the figures. A phenylalanine concentration of 0.9 mM is typical of that in the plasma of phenylketonuria patients, and a phenylpyruvic acid concentration of 1.2 mM is greater than that in the plasma of phenylketonuria patients (<1 mM) (Bedin et al., 2000; Berti et al., 2001). For ERK inhibitor trials, cells were preincubated for 1 h with 10 µM of the ERK inhibitor II, FR180204 (Calbiochem, San Diego, CA), dissolved in DMSO.
Real-time PCR
Real-time PCR was performed using the SYGR Green system (Applied Biosystems, Warrington, UK) as previously described (Park et al., 2009). GAPDH was used as the endogenous control gene. The relative between-group expression of the Ttr gene was calculated as 2-ΔΔCt (Livak and Schmittgen, 2001).
Reporter plasmid construct
A 356 bp mouse Ttr gene promoter containing BglII and HindIII site adapters was subcloned into the corresponding restriction sites of the luciferase reporter pGL3-basic vector (Promega, San Luis Obispo, CA). The subcloned sequences were verified by DNA sequence analyses.
Reporter gene transfection assays: luciferase assay
The luciferase assay was carried out using the Dual-Luciferase Reporter assay system (Promega, San Luis Obispo, CA) as previously described (Wang and Burker, 2007). Reporter plasmids containing the Ttr promoter region were transfected into subconfluent HepG2 cells using FuGENE HD transfection reagent (Roche Applied Science, Indianapolis, IN). To normalize the transfection efficiency, the pRL-cytomegalovirus promoter (Renilla luciferase vector, Promega) was cotransfected into each well. Each well of a 24-well culture plate received 1 µg of DNA mixture consisting of firefly reporters and Renilla control vector in the ratio of 40:1. After 24 h, each cell was treated with phenylalanine, phenylpyruvic acid, PBS, or DMSO. Cells were lysed using Promega lysis buffer. Firefly and Renilla luciferase activities were measured sequentially in the same sample using a Veritas microplate luminometer (Turner BioSystems, Sunnyvale, CA). Normalized luciferase activities were calculated by dividing the firefly luciferase activity by the Renilla luciferase activity. Relative luciferase activity is expressed as a percentage of the reporter activity relative to that of control cells.
Western blot analysis
Nuclear extracts and lysates were prepared as previously described (Wadman et al., 1997; Park et al., 2009). Western blotting was performed according to standard protocols using antibodies against HNF1, HNF3α/β, HNF4α, C/EBPα, C/EBPβ, histone H1 (Santa Cruz Biotechnology, CA), phospho-p38 MAP kinase, total-p38 MAP kinase, phospho-ERK1/2, total-ERK1/2 (Cell Signaling Technology, Beverly, MA), transthyretin (Dako, Carpinteria, CA), and GAPDH (AbFrontier, Seoul, Korea) (Ju et al., 2009).
Gas chromatography-mass spectrometric analysis (GC/MS) measurements of phenylpyruvic acid
Cells treated with 0.9 mM phenylalanine or 1.2 mM phenylpyruvic acid were sonicated and acidified to pH 2. The acidified solution was supplemented with NaCl and extracted with ethyl acetate. Tropic acid (Sigma-Aldrich) was added as an internal standard. The organic phase was separated by centrifugation and transferred to a new tube. The dried samples were mixed with N-methyl-N-trimethylsilyltrifluoroacetamide (Pierce Chemical Co., IL). Finally, the samples were incubated for 30 min at 80℃, and 2 µl of the analyte was injected into an HP-5MS column (Agilent J & W Scientific, Folsom, CA) and analyzed using an Agilent 7890A gas chromatograph coupled with an Agilent 5975C inert MSD mass spectrometer (Agilent Technologies, Santa Clara, CA).
Acknowledgments
This study was supported by a Korean Health 21 R&D Project from the Ministry of Health and Welfare, Republic of Korea (A010384).
Abbreviations
- C/EBP
CCAAT/enhancer binding protein
- HNF
hepatocyte nuclear factors
- PAH
phenylalanine hydroxylase
- SAPK
stress-activated protein kinase
- TTR
transthyretin
- Ttr
transthyretin
References
- 1.Acosta PB, Yannicelli S, Marriage B, Steiner R, Gaffield B, Arnold G, Lewis V, Cho S, Berstein L, Parton P, Leslie N, Korson M. Protein status of infants with phenylketonuria undergoing nutrition management. J Am Coll Nutr. 1999;18:102–107. doi: 10.1080/07315724.1999.10718836. [DOI] [PubMed] [Google Scholar]
- 2.Arnold GL, Vladutiu CJ, Kirby RS, Blakely EM, Deluca JM. Protein insufficiency and linear growth restriction in phenylketonuria. J Pediatr. 2002;141:243–246. doi: 10.1067/mpd.2002.126455. [DOI] [PubMed] [Google Scholar]
- 3.Awad MM, Enslen H, Boylan JM, Davis RJ, Grupposo PA. Growth regulation via p38 mitogen-activated protein kinase in developing liver. J Biol Chem. 2000;275:38716–38721. doi: 10.1074/jbc.M008040200. [DOI] [PubMed] [Google Scholar]
- 4.Bedin M, Estrella CH, Duarte DV, Ponzi D, Dutra-Filho CS, Wyse AT, Wajner M, Wannmacher CM. Platelet Na+, K+-ATPase activity as a possible peripheral marker for the neurotoxic effects of phenylalanine in phenylketonuria. Metab Brain Dis. 2000;15:115–121. doi: 10.1007/BF02679978. [DOI] [PubMed] [Google Scholar]
- 5.Berti SL, Bonan CD, da Silva FL, Battastini AM, Sarkis JM, Wannmacher CM. Phenylalanine and phenylpyruvate inhibit ATP diphosphohydrolase from rat brain cortex. Int J Dev Neurosci. 2001;19:649–653. doi: 10.1016/s0736-5748(01)00048-x. [DOI] [PubMed] [Google Scholar]
- 6.Buxbaum JN, Ye Z, Reixach N, Friske L, Levy C, Das P, Golde T, Masliah E, Roberts AR, Bartfai T. Transthyretin protects Alzheimer's mice from the behavioral and biochemical effects of Abeta toxicity. Proc Natl Acad Sci USA. 2008;105:2681–2686. doi: 10.1073/pnas.0712197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi SH, Leight SN, Lee VM, Li T, Wong PC, Johnson JA, Saraiva MJ, Sisodia SS. Accelerated Abeta deposition in APPswe/PS1deltaE9 mice with hemizygous deletions of TTR (transthyretin) J Neurosci. 2007;27:7006–7010. doi: 10.1523/JNEUROSCI.1919-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costa PP, Figueira AS, Bravo FR. Amyloid fibril protein related to prealbumin in familial amyloidotic polyneuropathy. Proc Natl Acad Sci USA. 1978;75:4499–4503. doi: 10.1073/pnas.75.9.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costa RH, Grayson DR, Darnell JE., Jr Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and alpha 1-antitrypsin genes. Mol Cell Biol. 1989;9:1415–1425. doi: 10.1128/mcb.9.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa RH, Grayson DR. Site-directed mutagenesis of hepatocyte nuclear factor (HNF) binding sites in the mouse transthyretin (TTR) promoter reveal synergistic interactionswith its enhancer region. Nucleic Acids Res. 1991;19:4139–4145. doi: 10.1093/nar/19.15.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa RH, Lai E, Darnell JE., Jr Transcriptional control of the mouse prealbumin (transthyretin) gene: both promoter sequences and a distinct enhancer are cell specific. Mol Cell Biol. 1986;6:4697–4708. doi: 10.1128/mcb.6.12.4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Episkopou V, Maeda S, Nishiguchi S, Shimada K, Gaitanaris GA, Gottesman ME, Robertson EJ. Disruption of the TTR gene results in mice with depressed levels of plasma retinol and thyroid hormone. Proc Natl Acad Sci USA. 1993;90:2375–2379. doi: 10.1073/pnas.90.6.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman JM, Babiss LE, Clayton DF, Darnell JE., Jr Cellular promoters incorporated into the adenovirus genome: cell specificity of albumin and immunoglobulin expression. Mol Cell Biol. 1986;6:3791–3797. doi: 10.1128/mcb.6.11.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatzis P, Kyrmizi I, Talianidis I. Mitogen-activated protein kinase-mediated disruption of enhancer-promoter communication inhibits hepatocyte nuclear factor 4alpha expression. Mol Cell Biol. 2006;26:7017–7029. doi: 10.1128/MCB.00297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ju SM, Song HY, Lee JA, Lee SJ, Choi SY, Park J. Extracellular HIV-1 Tat up-regulates expression of matrix metalloproteinase-9 via a MAPK-NF-kappaB dependent pathway in human astrocytes. Exp Mol Med. 2009;41:86–93. doi: 10.3858/emm.2009.41.2.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuo CF, Xanthopoulos KG, Darnell JE., Jr Fetal and adult localization of C/EBP: evidence for combinatorial action of transcription factors in cell-specific gene expression. Development. 1990;109:473–481. doi: 10.1242/dev.109.2.473. [DOI] [PubMed] [Google Scholar]
- 17.Li T, Jahan A, Chiang JY. Bile acids and cytokines inhibit the human cholesterol 7 alpha-hydroxylase gene via the JNK/c-junpathway in human liver cells. Hepatology. 2006;43:1202–1210. doi: 10.1002/hep.21183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Martynyuk AE, Glushakov AV, Sumners C, Laipis PJ, Dennis DM, Seubert CN. Impaired glutamatergic synaptic transmission in the PKU brain. Mol Genet Metab. 2005;86(Suppl 1):S34–S42. doi: 10.1016/j.ymgme.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 20.McDonald JD, Bode VC, Dove WF, Shedlovsky A. Pahhph-5: a mouse mutant deficient in phenylalanine hydroxylase. Proc Natl Acad Sci USA. 1990;87:1965–1967. doi: 10.1073/pnas.87.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonald JD, Charlton CK. Characterization of mutations at the mouse phenylalanine hydroxylase locus. Genomics. 1997;39:402–405. doi: 10.1006/geno.1996.4508. [DOI] [PubMed] [Google Scholar]
- 22.Mendelson KG, Contois LR, Tevosian SG, Davis RJ, Paulson KE. Independent regulation of JNK/p38 mitogen-activated protein kinases by metabolic oxidative stress in the liver. Proc Natl Acad Sci USA. 1996;93:12908–12913. doi: 10.1073/pnas.93.23.12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monteiro FA, Sousa MM, Cardoso I, do Amaral JB, Guimarães A, Saraiva MJ. Activation of ERK1/2 MAP kinases in familial amyloidotic polyneuropathy. J Neurochem. 2006;97:151–161. doi: 10.1111/j.1471-4159.2006.03716.x. [DOI] [PubMed] [Google Scholar]
- 24.Park JW, Park ES, Choi EN, Park HY, Jung SC. Altered brain gene expression profiles associated with the pathogenesis of phenylketonuria in a mouse model. Clin Chim Acta. 2009;401:90–99. doi: 10.1016/j.cca.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 25.Samadani U, Costa RH. The transcriptional activator hepatocyte nuclear factor 6 regulates liver gene expression. Mol Cell Biol. 1996;16:6273–6284. doi: 10.1128/mcb.16.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarkissian CN, Scriver CR, Mamer OA. Measurement of phenyllactate, phenylacetate, and phenylpyruvate by negative ion chemical ionization ± gas chromatography/mass spectrometry in brain of mouse genetic models of phenylketonuria and non-phenylketonuria hyperphenylalaninemia. Anal Biochem. 2000;280:242–249. doi: 10.1006/abio.2000.4542. [DOI] [PubMed] [Google Scholar]
- 27.Scriver CR, Kaufman S. Hyperphenylalaninemia: Phenylalanine hydroxylase deficiency. In: Scriver CR, Kaufman, Eisensmith RC, Woo SLC, editors. The Metabolic and Molecular Bases of Inherited Disease. New York, NY: McGraw-Hill; 2001. pp. 1667–1724. [Google Scholar]
- 28.Shenton A, Wells FE, Addison GM. Prealbumin as an indicator of marginal malnutrition in treated phenylketonuria; a preliminary report. J Inherit Metab Dis. 1983;6(Suppl):109–110. [Google Scholar]
- 29.Surendran S, Rady PL, Szucs S, Michals-Matalon K, Tyring SK, Matalon R. High level of orexin A observed in the phenylketonuria mouse brain is due to the abnormal expression of prepro-orexin. Biochem Biophys Res Commun. 2004;317:522–526. doi: 10.1016/j.bbrc.2004.03.079. [DOI] [PubMed] [Google Scholar]
- 30.Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, Burker PA. Effects of hepatocyte nuclear factor-4alpha on the regulation of the hepatic acute phase response. J Mol Biol. 2007;371:323–335. doi: 10.1016/j.jmb.2007.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Z, Tavares-Sanchez OL, Li Q, Fernando J, Rodriguez CM, Studer EJ, Pandak WM, Hylemon PB, Gil G. Activation of bile acid biosynthesis by the p38 mitogen-activated protein kinase (MAPK): hepatocyte nuclear factor-4alpha phosphorylation by the p38 MAPK is required for cholesterol 7alpha-hydroxylase expression. J Biol Chem. 2007;282:24607–24614. doi: 10.1074/jbc.M611481200. [DOI] [PMC free article] [PubMed] [Google Scholar]