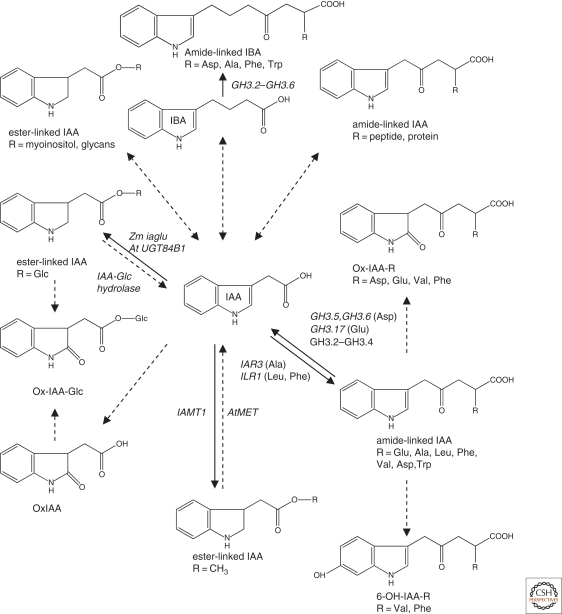
Figure 3.
IAA metabolism in Arabidopsis. Updated from Woodward and Bartel 2005. Genes identified since the Woodward and Bartel review include GS3.2-GS3.6 and GS3.17 (Staswick et al. 2002; Staswick et al. 2005). Recently identified IAA-metabolites include IAA-Leu, IAA-Ala (Kowalczyk and Sandberg 2001; Rampey et al. 2004), IAA-Phe, IAA-Val (Kai et al. 2007a), IAA-Trp (Staswick 2009), oxIAA-Asp, oxIAA-hexose (Ostin et al. 1998), oxIAA-Glu, -Val, -Phe and –glucose (Kai et al. 2007a), 6-OH-IAA-Val and -Phe (Kai et al. 2007a), and methyl-IAA (Qin et al. 2005). IBA is a substrate for some of the GH3 genes, as indicated by in vitro assay (Staswick et al. 2005). See Cohen and Bandurski 1982, Normanly 1997, and Seidel et al. 2006 for IAA metabolites identified in other species. Dashed arrows indicate that the enzyme activity has been observed in vitro only, or neither gene nor enzyme activity have been identified.