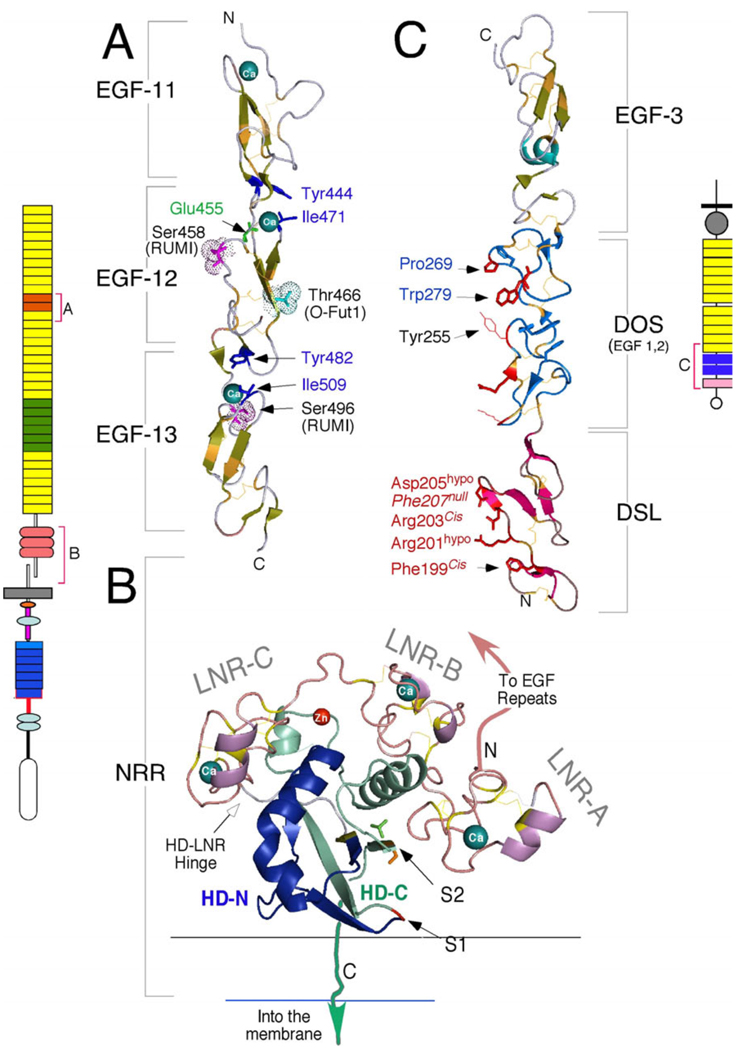
Figure 3.
Atomic resolution details of the ligand-binding domain (EGF 11–13) from human Notch1 (PDB:2VJ3), the NRR from human Notch2 (PDB:2OO4) and the Notch-binding domain from human Jagged1 (PDB:2VJ2) generated with MacPymol (http://www.pymol.org). A) The ligand-binding domain of Notch1 (schematically depicted to the left) is centered on EGF repeat 12. The essential amino acids that coordinate Ca++ binding are shown in blue. O-glycosylation (Ser458, Ser496) and O-fucosylation (Thr466) sites are shown. Of note, the equivalent mutation to Glu455Val abolishes ligand binding in Drosophila, and this interface in hNotch1 was suggested to interact with hJagged1 DSL based on in silico docking models (Cordle et al., 2008a). However, glycosylation on Ser458 may block access to this site. Thr466 is essential for productive Notch activation in the mouse but not in the fly. B) The NRR domain folds to protect the S2 cleavage site (colorized side chains), which is located in a pocket protected by LNR-A, the HD-C helix, and the LNR-B/A linker. The furin cleavage site S1 lies within an unstructured loop that was removed to facilitate crystallization. LNR repeats bind calcium; chelation of these Ca++ atoms lead to NRR dissociation and Notch activation. See text and (Gordon et al., 2007) for further details. C) The crystal structure of the DSL, DOS and EGF repeat 3 of hJagged1 (schematically depicted to the right in trans binding orientation), highlighting the putative Notch binding interface (facing left)). The DSL fold is distinct from the EGF fold; amino acids in DSL that were shown to be required for interaction with Notch are labeled in red (see (Cordle et al., 2008a) for detail). Phe207Ala substitution generates a null protein whereas Arg203Ala and Phe199Ala substitutions ablate trans but not cis binding. Asp205Ala and Arg201Ala are hypomorphic. The DOS domain contains two conserved, atypical EGF repeats (defined by the presence of the conserved amino acids shown in blue (Komatsu et al., 2008)). Tyr255 is characteristic of Jagged DSL ligands and is replaced by a small hydrophobic amino acid in Delta-like proteins; this residue may be involved in defining sensitivity to Fringe.