Figure 7.
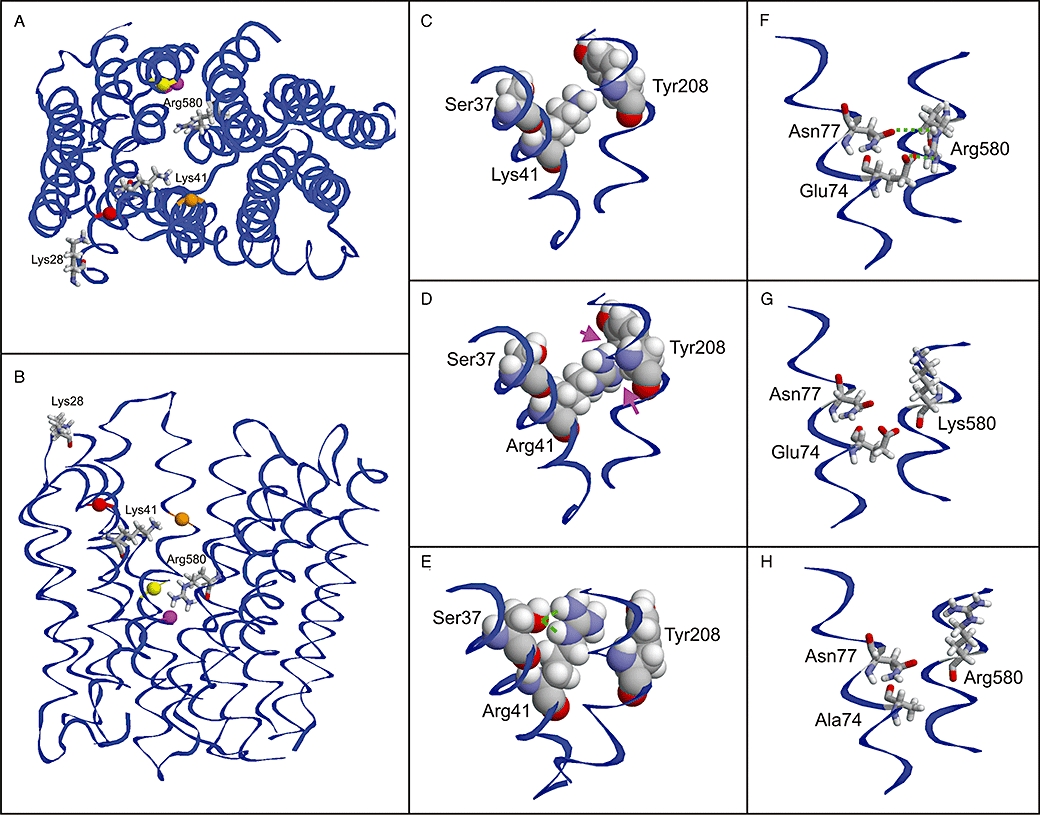
Structural modelling of OATP1B3. Top view (A) and side view (B) of the model showing the positions of the investigated residues. The positively charged amino acids are shown as stick presentation and are labelled. Adjacent amino acids, which are discussed in the text, are shown as balls and are colour coded as follows: Ser37 (red), Tyr208 (orange), Glu74 (magenta) and Asn77 (yellow). (C–E) Effect of the Lys41>Arg mutation: While the Lys41 side chain shows no interactions with adjacent amino acids (C), Arg41 exhibits minor clashes (D; magenta arrows), which probably lead to an alternative side chain conformation (E; green lines indicate an interaction with Ser37). (F–H) Effect of the Arg580>Lys and Glu74>Ala mutations: The orientation of Arg580 is stabilized by interactions with Glu74 and Asn77 (F), which cannot be formed by a lysine (G) resulting in an altered side chain orientation. The same loss of interactions is predicted for a Glu74>Ala mutation (H).
