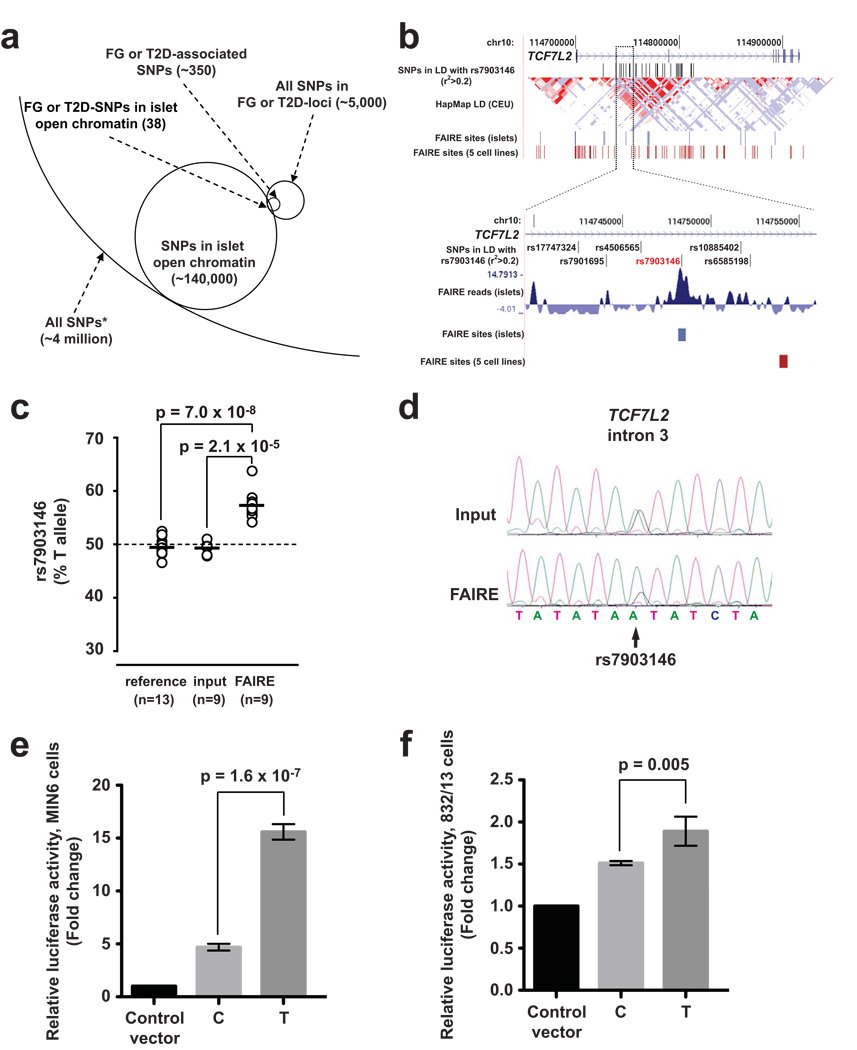
Figure 4. Allele-specific open chromatin and enhancer activity at the TCF7L2 locus.
(a) Schematic representation of how FAIRE-seq enables the identification of human sequence variants located in islet open chromatin. From ~4 million SNPs present in dbSNP with average heterozygosity >1%, 38 SNPs associated with T2D or fasting glycemia mapped to islet open chromatin sites. The analysis was carried out with all SNPs in strong linkage disequilibrium (r2>0.8) with an FG- or T2D-associated variant, which are labeled as FG or T2D SNPs, and FAIRE-seq sites identified with a liberal threshold. (b) Among TCF7L2 variants in linkage disequilibrium with rs7903146 (r2>0.2, top panel), only rs7903146 maps to an islet-selective FAIRE site. (c) In all 9 human islet samples that were heterozygous for rs7903146, the risk allele T was more abundant than the non-risk C allele in the open chromatin fraction, in contrast to input DNA or gDNA from unrelated heterozygous individuals. (d) Allelic imbalance for open chromatin at rs7903146 was verified in independent assays using quantitative Sanger sequencing (see also Supplementary Fig. 4b). (e) The risk allele T of rs7903146 exhibits greater enhancer activity than the non-risk allele C in MIN6 cells and (f) 832/13 cells. Standard deviations represent four independent clones for each allele. Results for inserts in the reverse direction are provided in Supplementary Fig. 4. P-values were calculated by two-sided t-test.