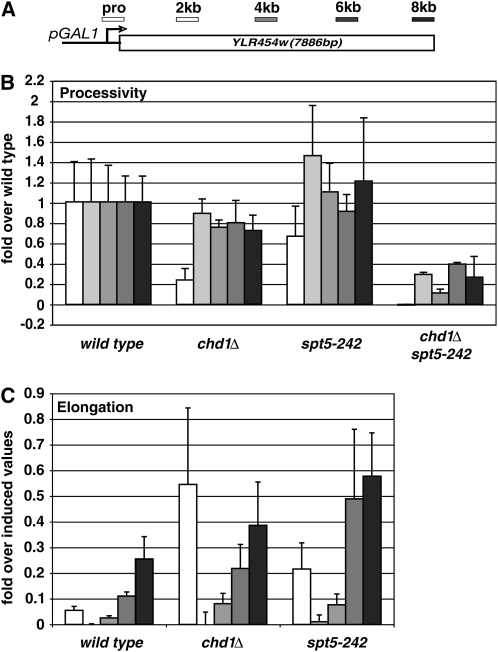
Figure 1.—
Reduced rate of RNA polymerase II elongation in spt5-242 mutant. (A) Location of ChIP probes on hybrid GAL1-YLR454W gene. (B) Measurement of RNAPII processivity. ChIP of RNAPII across the hybrid GAL1-YLR454W gene in wild-type cells and the indicated mutants was performed under inducing conditions. For each mutant, IP/Input values for each ChIP probe were determined and normalized to the corresponding value measured in the wild-type strain. (C) Measurement of elongation rate. Galactose-induced cells were treated with glucose to repress transcription from the GAL1-YLR454W gene and samples were processed for ChIP 5 min later. IP/Input values for each probe are expressed relative to the corresponding value for that probe measure just prior to addition of glucose.