Figure 2.
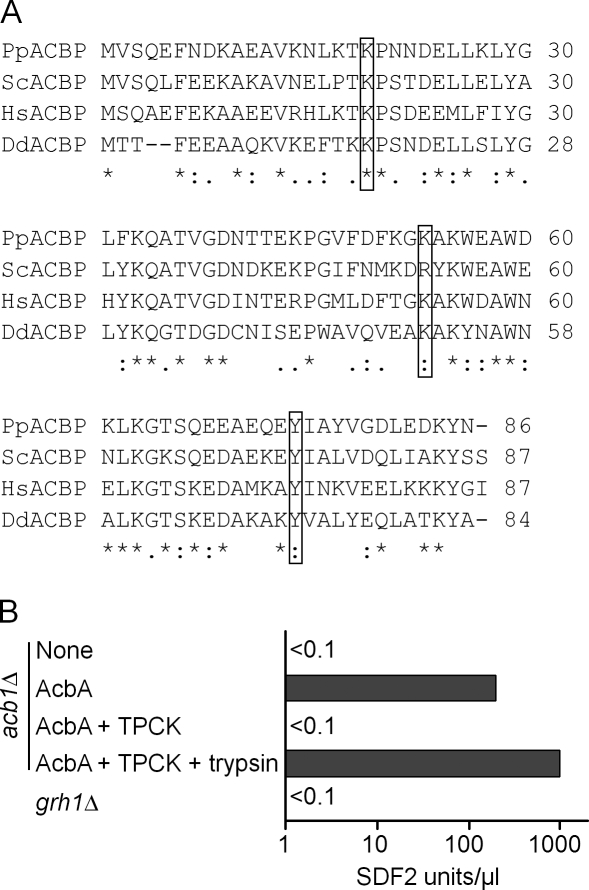
Primary sequence of P. pastoris Acb1 and activity of a P. pastoris protease that processes AcbA to SDF-2. (A) The primary sequences of Acbps from P. pastoris (NCBI Protein database accession no. XP_002490495), S. cerevisiae (NCBI Protein database accession no. NP_011551), Homo sapiens (NCBI Protein database accession no. NP_001073331), and D. discoideum (NCBI Protein database accession no. XP_646321) were aligned in CLUSTALW using the default alignment parameters (*, conserved residues). Putative, conserved trypsin-like cleavage site residues (K) are highlighted in boxed regions. The MatGAT program was used to calculate percent identity (I) and similarity (S) of the P. pastoris Acb1 protein with that of S. cerevisiae (I = 63.2%; S = 77%), H. sapiens (I = 55.2%; S = 77%), and D. discoideum (I = 45.3%; S = 62.8%). (B) Trypsin-like protease activity of acb1Δ cells. The acb1Δ cells were incubated in nitrogen starvation medium for 4 h without any addition (none) or in the presence of 100 pM recombinant AcbA, and extracellular media were collected, purified, and assayed for SDF-2–like activity (AcbA). 1 µM TPCK inhibitor was added 3 h after transfer to starvation medium and assayed (AcbA + TPCK). Part of the sample was digested with trypsin (see Materials and methods), purified, and assayed using the bioassay (AcbA + TPCK + trypsin). The GRASP-null mutant (grh1Δ) was also processed and tested for SDF-2–like activity like the other samples. Experiments were performed three times.