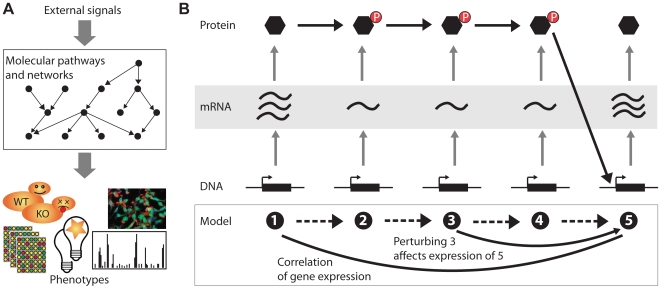
Figure 1. Cellular networks underlying observable phenotypes.
(A) Phenotypes are the response of the cell to external signals mediated by cellular networks and pathways. The goal of computation is to reconstruct these networks from the observed phenotypes. (B) Global molecular phenotypes like gene expression allow a view inside the cell but also have limitations. This is exemplified here in a cartoon pathway adapted from [61] showing a cascade of five genes/proteins (1–5). Proteins 1–3 form a kinase cascade, 4 is a transcription factor acting on 5. Up-regulation of 1 starts information flow in the cascade and results in 5 being turned on. In gene expression data this is visible as a correlation between 1 and 5 (represented as an undirected edge in the model). Experimentally perturbing a gene, say 3, removes the corresponding protein from the cascade, breaks the information flow, and results in an expression change at 5 (represented as an arrow in the model). However, the different phosphorylation and activation states of proteins 2–4 will most probably not be visible as changes in gene expression. Thus, because of the pathway mostly acting on the protein level most parts of the cascade (dashed arrows in the model) can not be inferred from gene expression data directly.