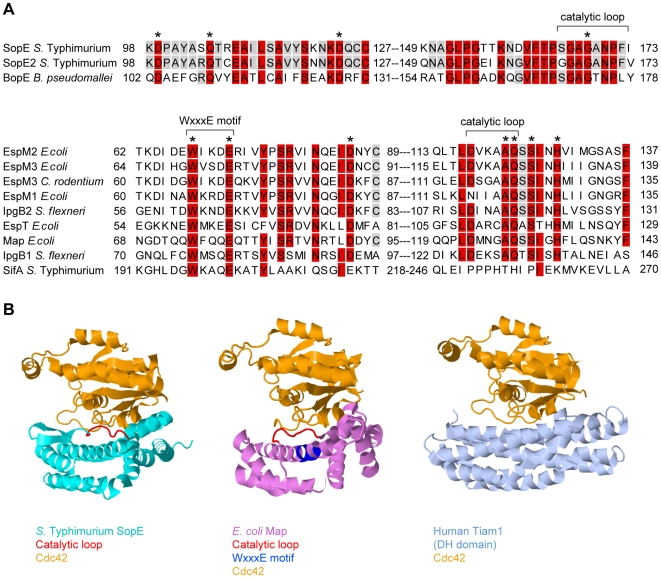
Figure 2. Sequence and structural comparison of GEFs from different origins.
(A) Sequence alignment of SopE-type (upper alignment) and WxxxE-type (lower alignment) bacterial effector proteins with postulated Rho GTPase activity; >80% identity is shaded in red, >50% in grey. The residues shown to be important for catalytic GEF activity of SopE [13] or Map [19] are marked with asterisks. WxxxE motif and catalytic loops highlighted in (B) are marked with brackets. Note that SifA has a completely different amino acid sequence in its catalytic loop compared to the other proteins in the WxxxE family [19]. (B) Crystal structures of GEFs from different organisms in complex with Cdc42 (orange). Left panel: SopE (cyan) from S. Typhimurium [12]; the catalytic loop is highlighted in red. Middle panel: Map (pink) from enteropathogenic E. coli [19]; the WxxxE motif is highlighted in blue and the catalytic loop in red. Right panel: Human Tiam1 (pale blue); only the Rho GTPase binding DH domain is shown [14]. Structures were created using Jmol (http://www.jmol.org/).