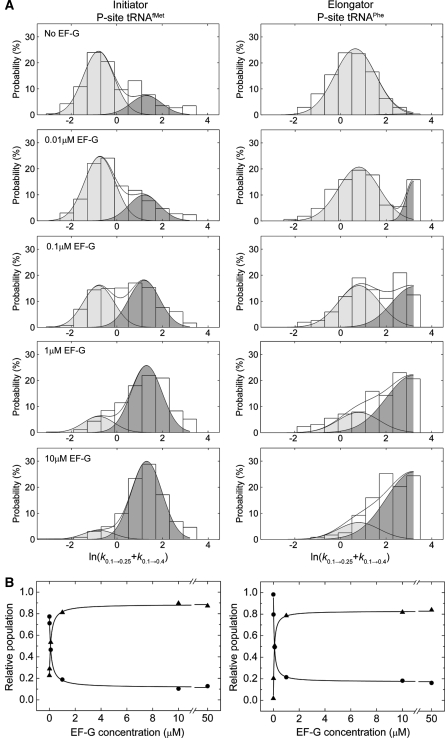
Figure 5.
EF-G-mediated dynamic mode switching in the ribosome reveals the existence of two distinct low-FRET states. (A) Histograms indicating the distributions of the logarithm of the transition rate out of the low-FRET state (klow →=k0.1 → 0.25+k0.1 → 0.4), calculated for each individual molecule (see Materials and methods section), as a function of EF-G concentration. At each concentration, including in the absence of EF-G, a bimodal distribution of rates is observed that represent slow (light grey) and fast (dark grey) dynamic modes of the ribosome. The populations in each mode were estimated by fitting the distributions to a double Gaussian function (R2>0.9) with mean values μ=−0.8 (s.d., σ=1.0; klow →slow≈0.5 per s) and μ=1.4 (σ=1.0; klow →fast≈4 per s) for the tRNAfMet-bound complex (left panels), and μ=0.8 (σ=1.2; klow →slow≈2 per s) and μ=3.2 (σ=1.2; klow →fast⩾25 per s) for the tRNAPhe-bound complex (right panels). In the case of tRNAPhe, only a portion of the fast dynamic mode distribution was observed due to the short low-FRET state lifetime relative to the experimental time resolution. (B) Plots showing the change in integrated areas of the slow and fast dynamic modes for each system as a function of EF-G concentration. Fitting changes in integrated areas of slow and fast populations to hyperbolic functions provides an estimate of the apparent KD (∼80 nM) for EF-G(GDPNP) binding to the ribosome (R2>0.9).