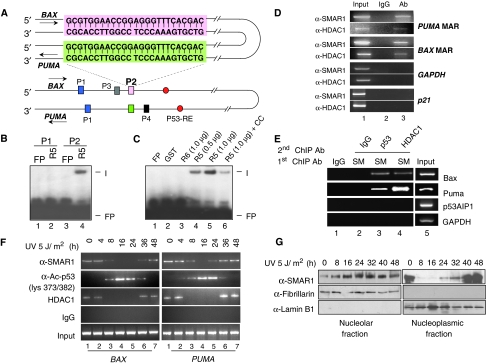
Figure 4.
SMAR1 regulates BAX and PUMA through MAR. (A) Schematic representation of BAX and PUMA promoter aligned with complementary strands showing the locations of two identical sequences P1 (blue) and P2 (pink in BAX and green in PUMA) in proximity to p53 response element (red). The sequence of P2 is boxed. (B) EMSA showing specific binding of SMAR1 (R5) to probe P2 (lane 4), but not to probe P1. (C) Purified GST (lane 2), R6 (lane 3) and R5 in increasing doses (lanes 4 and 5) were incubated with probe P2. Formation of complex (I) was visualized by autoradiography. Binding specificity of the complex in the presence of 10-fold molar excess of cold competitor (cold probe P2) is shown in lane 6. Free probe is denoted as FP. (D) Chromatin from HCT116 p53+/+ cells was immunoprecipitated with SMAR1 and HDAC1 antibodies (lane 3). PCR amplification was performed on MAR regions of BAX and PUMA. GAPDH and p21 promoters were used as control. Parallel immunoprecipitation with control IgG antibody is shown in lane 2. Lane 1 denotes input control. (E) Sequential ChIP on BAX and PUMA promoter MARs using anti-SMAR1/p53 (lane 3) and anti-SMAR1/HDAC1 (lane 4) antibodies in nuclear matrix fraction of HCT116 cells. Lanes 1 and 5 shows IgG and input control, respectively. (F) ChIP showing occupancy of SMAR1 on BAX (left panel) and PUMA (right panel) promoter on low-dose UV irradiation. Cross-linked chromatins from UV-irradiated HCT116 p53+/+ cells were pulled with SMAR1, p53, acetylated p53 lys373/382 and HDAC1 antibodies and bound chromatin fragments were detected by PCR. (G) Western blotting of SMAR1 in nucleolar and nucleoplasmic fraction of UV-irradiated (5 J/m2) HCT116 p53+/+ cells at different time points.