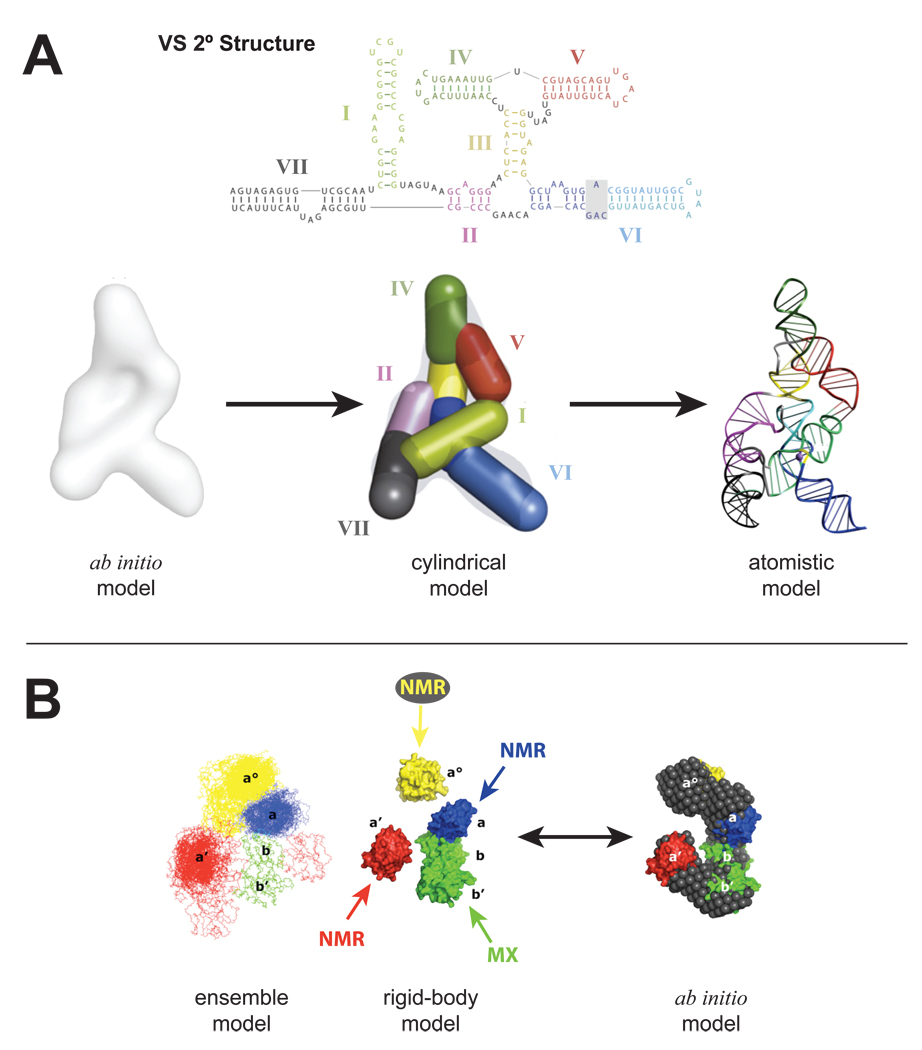
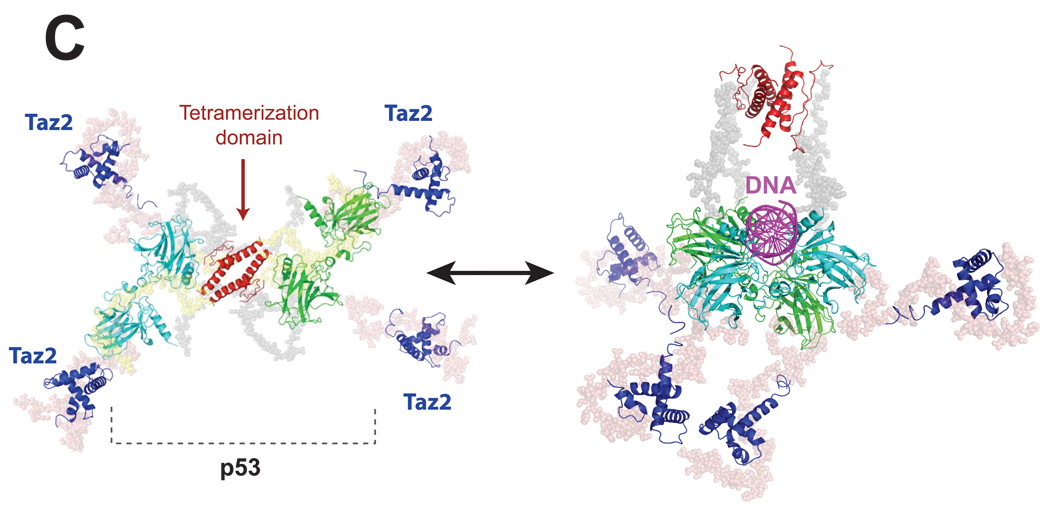
Figure 3.
SAXS extends modeling of most large multi-domain macromolecular machines and suggests that flexibility and unstructured regions are generally critical for function. A. Construction of the VS ribozyme solution structure [21••] started with the ab initio model which was used to place cylindrical elements that corresponded to the helical elements described by its secondary structure. The resulting cylindrical model was then converted to a residue specific model and subject to cycles of energy minimization refinement to produce the final atomistic solution model of the entire VS ribozyme. B. The entire ERp72 solution structure [22] was achieved by refining the SAXS data of ERp72 against a starting model composed of its individual domains that were previously determined by a combination of NMR and MX. 380 separate rigid-body modeling runs converged to show a C-shape ensemble consistent with a separate ab initio reconstruction. C. The full-length p53-Taz2-DNA complex (right) [31••] was modeled from SAXS data of the all protein complex (left) [32]. The core (cyan and green) and tetramerization (red) domains were determined by MX and Taz2 was determined by NMR. The three high-resolution structures were incorporated into a rigid body analysis to produce the final cross-shaped structure showing fully extended arms held together by the tetramerization domain. Flexible regions were modeled with dummy residues (gray beads). Additional SAXS data collected in the presence of DNA (magenta) was then subject to additional rigid body modeling using an MX derived structure of the core domain bound to DNA. The binding of Taz2 to p53 and the associated p53-DNA complex facilitated visualization of the N-terminal regions of p53, an observation not seen in a previous EM study.