Figure 2. Justaposition of RRMs in PTB1:34 increased RNA binding affinity.
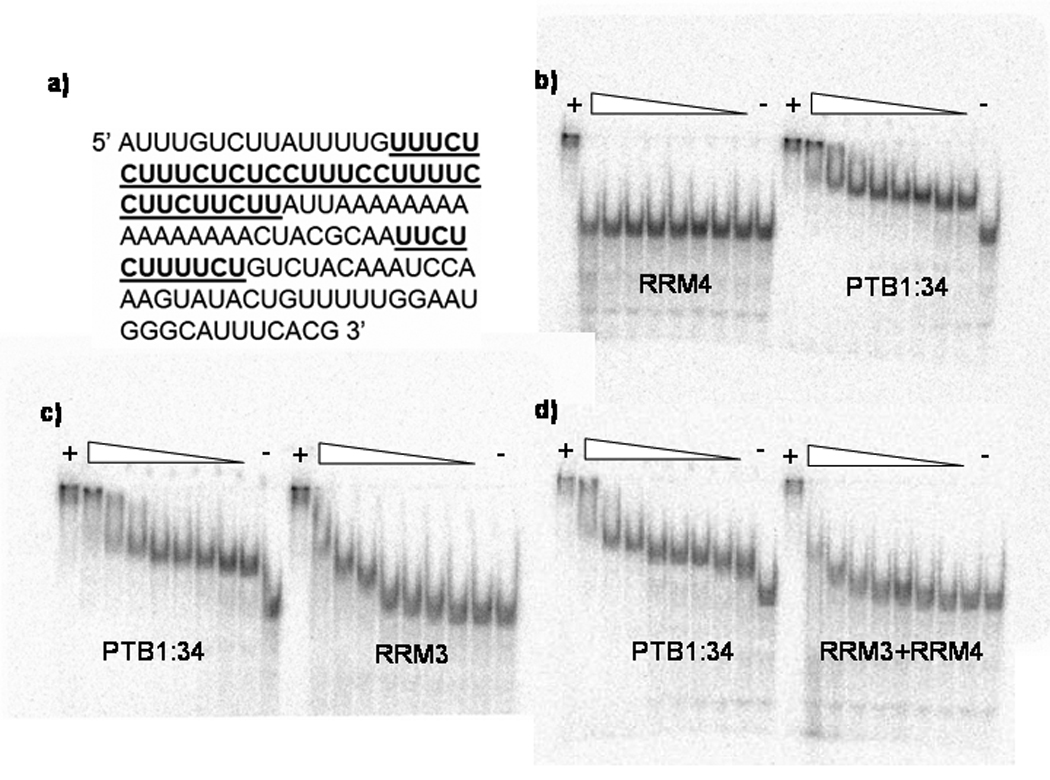
Electrophoretic mobility shift assays were used to compare the relative binding affinities of the GABAA γ2 pre-mRNA intron (a) to the PTB1:34 protein constructs. PTB1:34 binds at the lowest protein concentration tested, 10 nM, while RRM4 does not bind at all, even at the highest concentration tested, 10 µM (b). A similar comparison in (c) shows that RRM3 does bind to this RNA, but with around 50-fold lower affinity than PTB1:34, as the first significant band shift does not occur at protein concentrations less than 500 nM. Mixing RRM3 and RRM4 does not rescue the RNA binding (d), since an equimolar mixture of the two domains binds with affinity similar to that of RRM3 alone. All EMSAs were run at 4° C, and included a lane with RNA only as a negative control, and a lane with 800 nM full-length PTB, which is known to bind to this RNA with high affinity (KD ~ 1 nM for the first binding event in these solution conditions), as a positive control.
