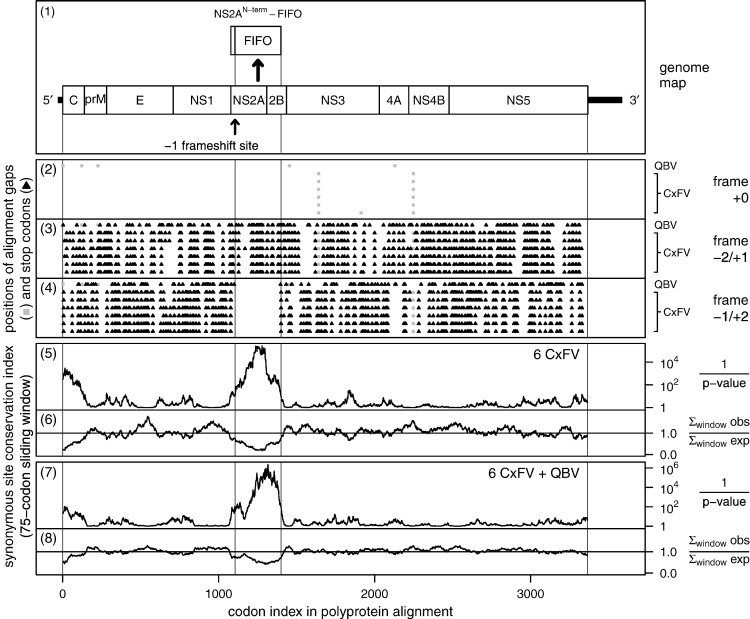
Fig. 2.
Conservation at synonymous sites in CxFV and QBV. Conservation was calculated for an input alignment comprising the polyprotein coding sequences from the CxFV sequences AB262759, AB377213, FJ663034, FJ502995, GQ165808 and EU879060, and the QBV sequence FJ644291. Panel 1 shows a map of the CxFV genome, along with the proposed overlapping gene fifo and the proposed frameshift site. Panels 2–4 show the positions of stop codons in the three forward reading frames. The + 0 frame is the polyprotein frame and is therefore devoid of stop codons. Note the conserved absence of stop codons in the − 1/+ 2 frame within the fifo ORF. Panels 5–8 depict the conservation at polyprotein-frame synonymous sites. Panels 5 and 7 depict the probability that the degree of conservation within a given window could be obtained under a null model of neutral evolution at synonymous sites, while panels 6 and 8 depict the absolute amount of conservation as represented by the ratio of the observed number of substitutions within a given window to the number expected under the null model (see Firth and Atkins, 2009, for details). Note that the large sliding window size (75 codons) is responsible for the broad smoothing of the conservation scores at either end of the fifo ORF. The range of features detected by this type of analysis depends partly on the sliding window size − 75 codons is optimal for features with a width of 75 codons. If features cover fewer codons (e.g., many non-coding functional elements), then the corresponding conservation peaks may be diluted to such an extent that they are not distinguishable from background noise.