Figure 1.
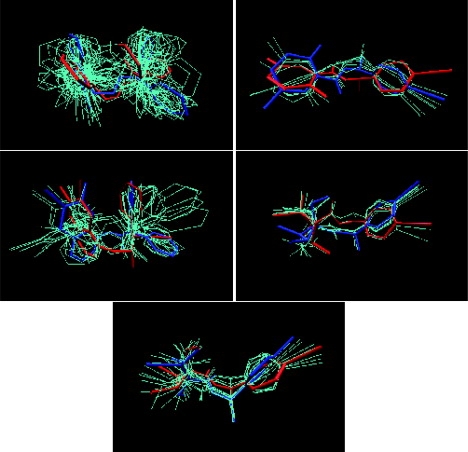
Superimposition of conformations for ligands to scytalone dehydratase. The blue conformer is the most favorable in the free state (that is, the reference conformer for the well with the largest calculated configuration integral). The red conformer is that of the ligand in the bound state, from the crystal structure. The remaining cyan structures are other suboptimal low-energy conformers in the free state. Only heavy atoms are displayed. The five ligands shown are (3-aminomethyl-cinnolin-4-YL)-(3,3-diphenyl-allylidene)-amine (PDB code 3std), N-[1-(4-bromophenyl)ethyl]-5-fluoro salicylamide (PDB code 4std), 6,7-difluoro-quinazolin-4-YL)-(1-metyl-2,2-diphenyl-ethyl)-amide (PDB code 5std), 2,2-difluoro-1-methanesulfinyl-3-methyl-cyclopropanecarboxylic acid [1-(4-bromo-phenyl)-ethyl]-amide (PDBcode 6 std), and (1RS,3SR)-2,2-dichloro-N-[(R)-1-(4-chlorophenyl)ethyl]-1-ethyl-3-methylcyclopropanecarboxamid (PDB code 7std).
