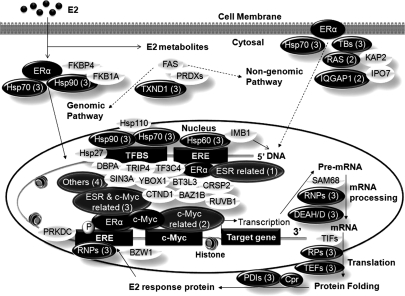
Fig. 7.
Proposed ERE complex consists of proteins classified in Table I as only ERα-related (1), only c-Myc-related (2), both ERα- and c-Myc-related (3), and others (4) and other identified proteins. Proteins in black ovals are significantly affected by E2 stimulation, and proteins in open ovals are transcription factors identified from the pulldown but without significant changes induced by a 24-h E2 stimulation. PRDXs, peroxiredoxins PRDX1, PRDX2, PRDX3, and PRDX6; TBs, tubulin; RAS, Ras-related proteins; RNPs, ribonucleoprotein; TIFs, translation initiation factor; TEFs, translation elongation factor; RPs, ribosomal protein; PDIs, protein-disulfide isomerases; Cpr, chaperone; BAZ1B, bromodomain adjacent to zinc finger domain protein 1B; BT3L3, transcription factor BTF3 homolog 3; CTND1, catenin δ-1; TRIP4, activating signal cointegrator 1; CRSP2, cofactor required for Sp1 transcriptional activation subunit 2; DBPA, DNA-binding protein A; RUVB1, RuvB-like 1; SIN3A, paired amphipathic helix protein Sin3a; TF3C4, general transcription factor 3C polypeptide 4; YBOX1, nuclease-sensitive element-binding protein 1; TFBS, transcription factor binding sites; ESR, ERα; FAS, fatty acid synthase; PRKDC, DNA-dependent protein kinase catalytic subunit.