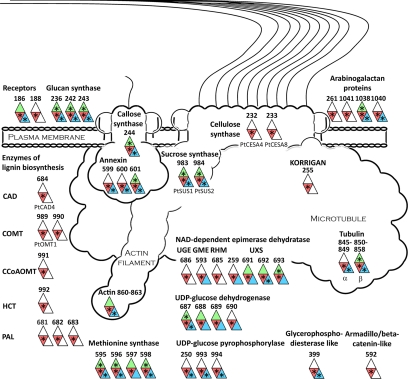
Fig. 6.
Proteins involved in wood formation. A schematic model of the cellulose-synthesizing complex and other proteins associated with wood formation, such as enzymes involved in lignin biosynthesis, is shown. The numbers outside each symbol refer to the gene model ID in Table I for integral proteins and supplemental Table 3 for soluble proteins. Only proteins detected in the xylem plasma membranes are included, and the color code is green for leaf, red for xylem, and blue for cambium/phloem plasma membranes. A star in a colored field indicates that the protein is classified as top rank in that tissue. When available, Populus names are stated below the symbol. Abbreviations for the NAD-dependent epimerase dehydratase family (11) are: UGE, UDP-d-glucose 4-epimerase; GME, GDP-d-mannose 3,5-epimerase; RHM, UDP-l-rhamnose synthase; UXS, UDP-d-apiose/xylose synthase. Abbreviations for the lignin enzymes are: CAD, cinnamyl alcohol dehydrogenase; COMT, caffeic acid O-methyltransferase; CCoAOMT, caffeoyl-CoA 3-O-methyltransferase; HCT, hydroxycinnamoyltransferase; PAL, phenylalanine ammonia-lyase.