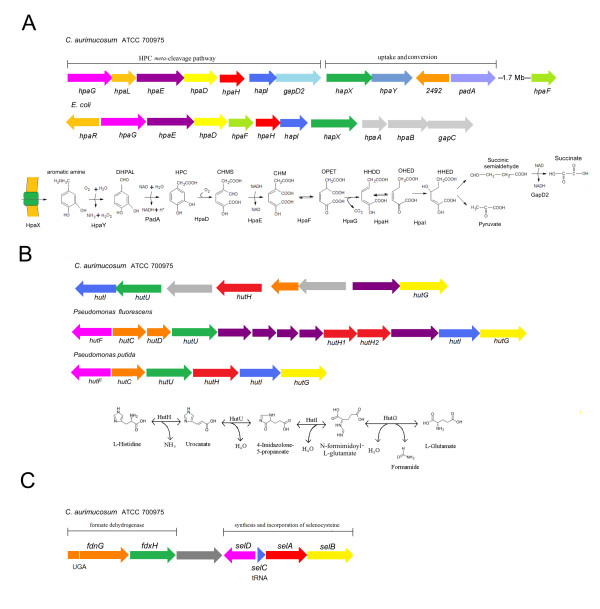
Figure 5.
Specific features of the C. aurimucosum ATCC 700975 metabolism detected by the analysis of singletons. (A), Genes and pathway for the catabolism of aromatic amines. Genetic maps of the hpa gene clusters encoding the HPC meta-cleavage pathway in C. aurimucosum and E. coli are presented. Genes with similar colour participate in the same enzymatic step of the pathway. The arrows show the direction of gene transcription. Abbreviations in the biochemistry of the pathway: DHPAL, 3,4-dihydroxyphenylacetaldehyde; HPC, homoprotocatechuate; CHMS, 5-carboxymethyl-2-hydroxy-muconic semialdehyde; CHM, 5-carboxymethyl-2-hydroxy-muconic acid; OPET, 5-oxo-pent-3-ene-1,2,5-tricarboxylic acid; HHDD, 2-hydroxy-hept-2,4-diene-1,7-dioic acid; OHED, 2-oxo-hept-3-ene-1,7-dioic acid; HHED, 2,4-dihydroxy-hept-2-ene-1,7-dioic acid. The enzymes are: HpaX, transport protein; HpaY, monoamine oxidase; PadA, phenylacetaldehyde dehydrogenase; HpaD, HPC 2,3-dioxygenase; HpaE, CHMS dehydrogenase; HpaF, CHM isomerase; HpaG, OPET decarboxylase; HpaH, hydratase; HpaI, HHED aldolase; GabD2, succinic semialdehyde dehydrogenase. (B), Genes and pathway for the catabolism of L-histidine. Genetics maps of the hut gene clusters coding for histidine catabolism in C. aurimucosum and Pseudomonas species. Genes with similar colour participate in the same enzymatic step of the pathway. The enzymes of the hut pathway are: HutH, histidine ammonia-lyase; HutU, urocanate hydratase; HutI, imidazolonepropionase; HutG, formimidoylglutamase. (C), Chromosomal gene region for selenol formate dehydrogenase and selenocysteine synthesis and incorporation in C. aurimucosum ATCC 700975. The UGA (opal) stop used for recoding the fdnG gene is indicated. The selC gene encodes the specific tRNASec.