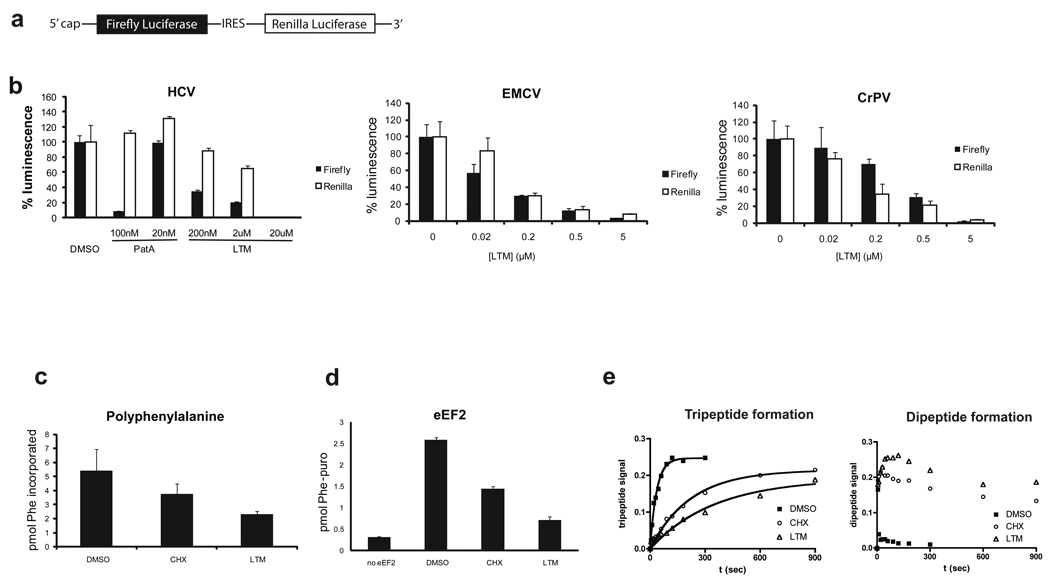
Figure 4. Effects of LTM and cycloheximide on different steps of translation elongation.
a. Configuration of the IRES reporters. Expression of firefly luciferase remains cap-dependent, while translation of renilla luciferase is under control of an IRES element. b. LTM inhibits IRES-mediated translation to a similar extent as cap-dependent translation. Pateamine A (PatA), which inhibits eIF4A-dependent translation initiation was chosen as a positive control. Error bars denote standard deviation. c. LTM inhibits poly-phenylalanine synthesis on a poly-uridine template. Phe-tRNA charged with [14C]phenylalanine was incubated with eEF1A, eEF2, ribosomes, poly(U) and GTP at 25°C for 2 min. Cycloheximide and LTM concentrations were both 200 µM. d. LTM inhibits eEF2-mediated translocation. Assay was performed as eEF1A assay, except for the use of GTP. After a 10-min preincubation, puromycin, indicated inhibitor, eEF2 and GTP were added. Formation of phenylalanyl puromycin was measured by scintillation counting of ethyl acetate extractable material. e. LTM and CHX decrease rate of tripeptide formation. The ability of pre-assembled initiation complexes to synthesize a tripeptide (Met-Phe-Phe) was measured over time. LTM and CHX treatments resulted in accumulation of didpeptides (right panel) and greatly reduced the rate of tripeptide formation (left panel). The measurements indicate the fraction of total input radioactivity. Bars in b–d represent s.d. from at least three repeats of each experiment.