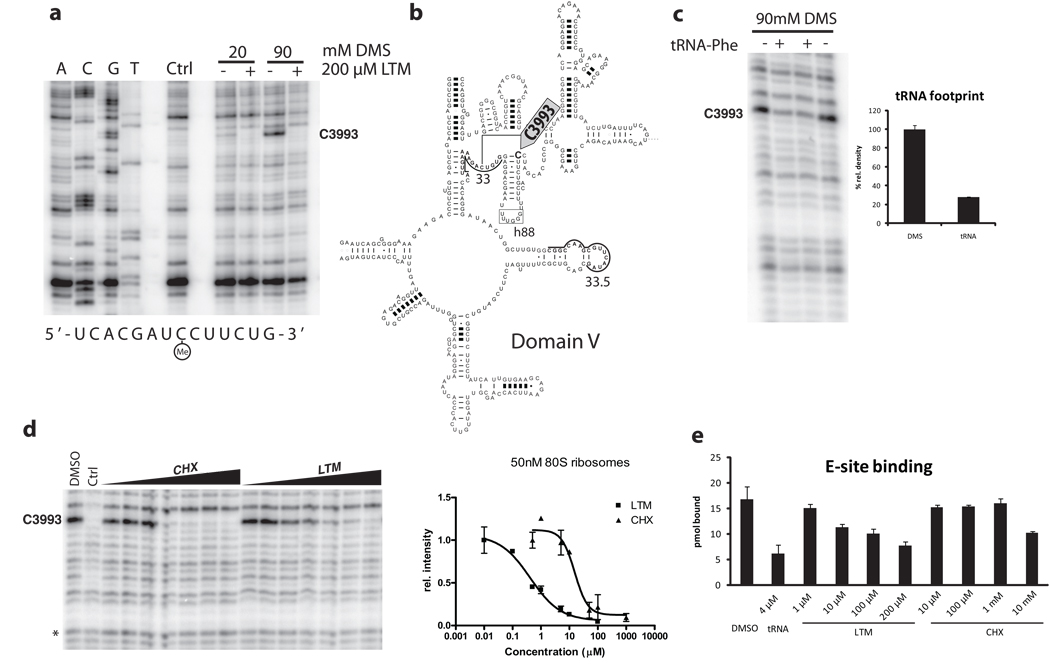
Figure 5. Footprinting analysis revealed the common binding sites of LTM and cycloheximide at the E-site of the larger ribosome subnit.
a. LTM binds to the 60S ribosomal exit site. 80S ribosomes were incubated with 200 µM LTM and methylated using 20 and 90 mM dimethyl sulfate. Extracted rRNA was hybridized to primer 33 or 33.5 (underlined) and reverse transcribed before electrophoresis. Ctrl denotes unmethylated rRNA. b. The binding site in domain V of the 28S rRNA at the base of hairpin 88 (arrow). c. The putative glutarimide-binding site coincides with the binding site of the 3’ end of deacylated tRNA at the E-site of the large ribosomal subunit. Deacylated Phe-tRNA was incubated with 80S ribosomes before DMS methylation and extraction. d. Both LTM and cycloheximide bind to the same site on the 60S ribosomal subunit in a dose-dependent manner. The KD values were estimated to be 500 nM for LTM and 15 µM for cycloheximide. e. LTM but not cycloheximide decrease binding of deacylated tRNA to the E-site. Ribosomes were incubated with [32P]-labeled deacylated Phe-tRNA in presence of LTM or cycloheximide at the indicated concentration. Excess cold tRNA was used as a positive control. Error bars denote standard deviation. Bars in c, d and e represent s.d..