Figure 1.
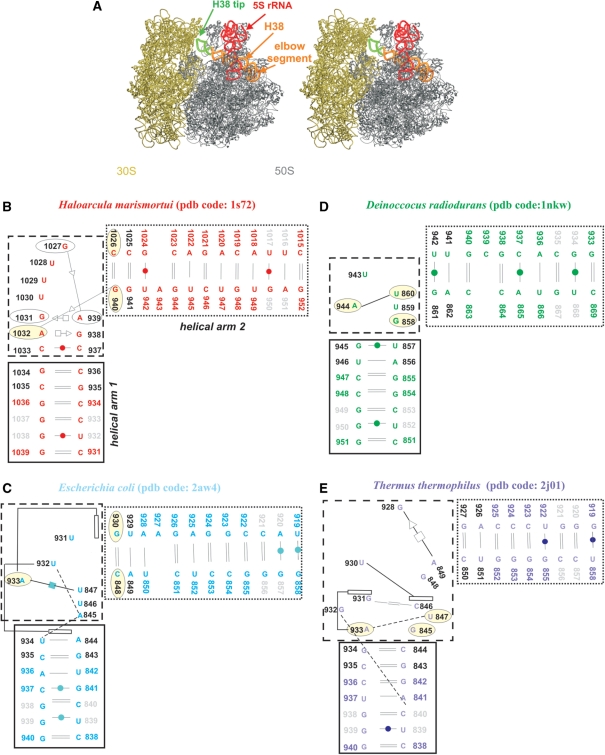
(A) Stereo view of E.c. 70S ribosome (50S is in grey; PDB code 2aw4 and 30S is in tan; PDB code 2avy) with highlighted 5S rRNA (red ribbon). The H38 (orange ribbon) was taken from E.c. 50S subunit (PDB code: 1pny) (68) where its complete structure is present, including its tip (green ribbon). The tip is missing in the atomic resolution structures. Positioning of H38 from 1pny structure into 2aw4 structure was carried out based on superposition of the 1pny and 2aw4 structures. (B–E) Secondary structures of the studied crystal H38 segments with base pairs marked according to the classification by Leontis and Westhof (62). The black and dotted boxes specify helical arms 1 and 2 while the dashed box indicates the central hinge segment which is defined as the area with the sharp bend of the backbone and the adjacent non-WC base pairs. The light yellow ellipses mark bases forming base triples in simulations (H.m. and T.t. structures have triples present already in the crystal structures). Dashed lines represent single H-bonds and empty rectangles mark stacking. Black residue numbers indicate fragments defining the α interhelical angle while grey residue numbers indicate fragments defining the β interhelical angle (the centers of mass of the two angles coincide in the central hinge segment; see Materials and Methods for the exact numbering). Note that the local interactions in the T.t. elbow segments are not identical in different X-ray structures (25,26) underscoring some uncertainties in the available experimental structures. The pairing suggested by the X-ray T.t. structure with PDB code 1vsp is shown in Supplementary Figure S2 and is actually more ordered and more consistent with the base pair classification by Leontis and Westhof. See the legend of Table 1 for explanation of the discontinuity of nucleotide numbering in the T.t. structure.