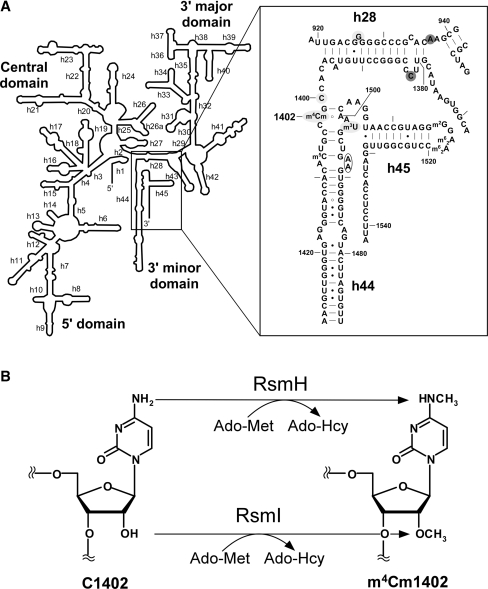
Figure 1.
The secondary structure of the decoding center in the E. coli 16S rRNA and the enzymatic formation of m4Cm1402. (A) The secondary structure of the E. coli 16S rRNA is shown on the left. Helix 44 and surrounding regions are shown on the right, with helix and base numbers indicated. A number of modified nucleosides including 5-methylcytidine (m5C), N4, 2′-O-dimethylcytidine (m4Cm) and 3-methyluridine (m3U) are also shown. The bases annotated as A-site (blank circle), P-site (shaded with light gray) and E-site (shaded with dark gray) are indicated (4,6). (B) Enzymatic formation of m4Cm1402. N4-methylation and 2′-O-methylation of C1402 are catalyzed by RsmH and RsmI, respectively, using Ado-Met as a methyl group donor. After the reaction, Ado-Met is converted to Ado-Hcy.