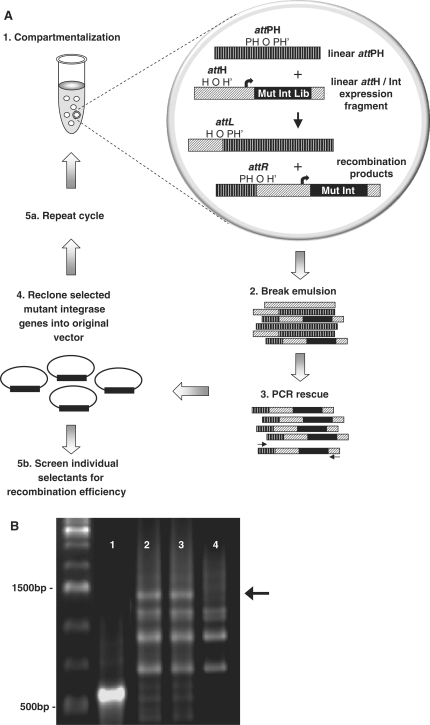
Figure 2.
Selection of novel integrase mutants by IVC. (A) Schematic of the IVC protocol for selecting novel integrase mutants. (1) A library of integrase mutants is constructed using error-prone PCR. Linear mutant integrase expression constructs with appended attH sites are segregated into the aqueous compartments of a water-in-oil emulsion, together with a separate linear attPH construct. Compartmentalization ensures that after translation, an active integrase mutant only recombines into the attH sequence appended to it, and not that of other integrase mutants. (2,3,4) The emulsion is disrupted, and genes encoding the active integrase mutants are amplified by PCR for characterization and further rounds of selection. PCR primers (arrows) only amplify recombination products containing the genes of integrase mutants capable of performing the attH × attPH recombination event. (B) PCR-rescue of integrase selectants generated by IVC. DNA encoding active integrase mutants was amplified by PCR after round 1 of the selection. Lanes 1 and 2: integrase mutant library plus attPH substrate. The primer pair used in Lane 1 produces a short ∼500 bp amplicon that does not contain the full-length integrase gene, while the pair used in lane 2 produces a longer ∼1400 bp amplicon that contains the full-length integrase gene. Lane 3: integrase mutant library plus attPH substrate and competing attB and attP substrates. Lane 4: negative control selection (identical to lane 3 but with inactive IVC extract). PCR amplification in lanes 2–4 was performed with the same primer pair.