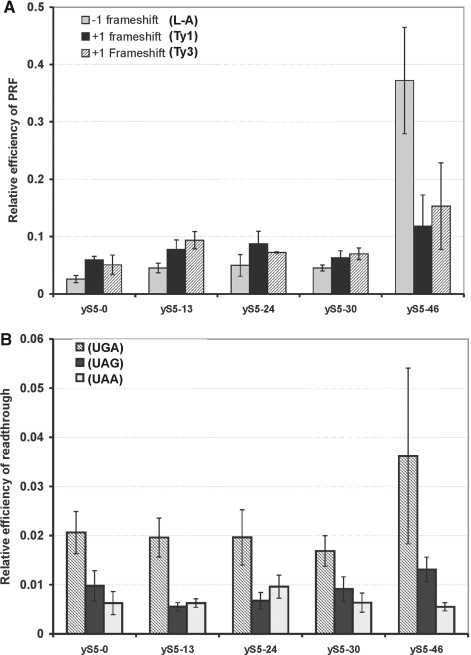
Figure 7.
Translation fidelity in the wt and mutant rpS5s yeast strains. (A) Programmed frameshifting efficiency. WT and mutant rpS5’s strain were transformed with the control, L-A, Ty1 and Ty3 frameshift reporter plasmids. Dual-luciferase assays were performed and programmed frameshifting (PRF) efficiencies were calculated as described (6,16,17,23). Mean efficiencies determined from at least six independent experiments are plotted and bars represent the corresponding standard error. (B) Termination codon read-through efficiency in wild-type and mutant yeast strains. The wt and human rpS5 mutant yeast strains expressing human rpS5 were transformed with control, UAA, UAG and UGA reporter plasmids and read-through efficiencies at each terminator were determined as described (6,14,15,21). Mean efficiencies determined from at least six independent experiments are plotted and bars represent the corresponding standard error. The significance of differences in signals between mutant and wt strain in all the experiments (P) above was at least <0.05, with the exception of UGA read-through signal in BY47yS5-46 strain.