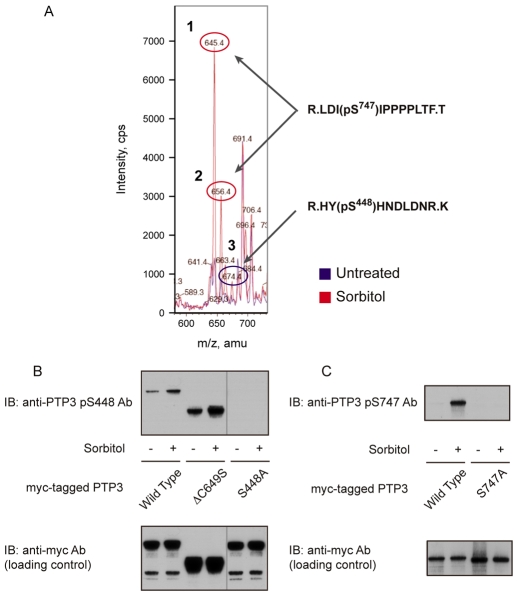
Fig. 1.
Identification of osmotic-stress-induced phosphorylation sites on PTP3. (A) Comparative mass spectrometry of the ΔCS form of myc-tagged PTP3 isolated from control and sorbitol-treated cells. Cells treated with either KK2 or KK2 containing sorbitol (200 mM) for 5 minutes were fractionated by anti-c-myc immunoprecipitation, followed by SDS gel electrophoresis. The myc:PTP3ΔCS bands were excised from the gel and digested with trypsin and chymotrypsin; because initial experiments showed that, to obtain a reasonable representation of the different parts of the protein, it was necessary to double digest in this way. With this enzyme combination, approximately 50% of residues proved analysable. The digested samples were analyzed with nano-liquid chromatography tandem mass spectrometry and the results were overlaid, with the untreated sample shown as a blue line and the sorbitol-treated sample shown in red. From the MS-MS analysis, peptides 1 and 2 were identified as LDI(pS)IPPPPLTF (phospho-Ser747) and peptide 3 as HY(pS)HNDLDNR (phospho-Ser448). (B) Analysis of phospho-Ser448- and phospho-Ser747-specific antibodies. Because of the low amount of PTP3 protein present in the cell, neither of the two antibodies could detect endogenous protein. Thus, PTP3-overepressing cells were used to analyze phosphorylation levels. Total cell lysate of PTP3-overexpressing cells was used for the phospho-Ser448 antibody and immunoprecipitated PTP3 protein was used for the phospho-Ser448 antibody.