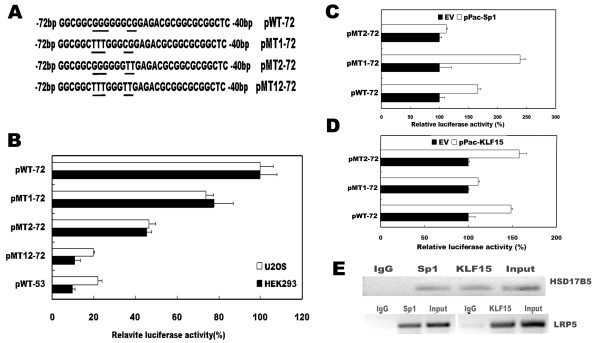
Figure 3.
KLF15 and Sp1 binding sites located between -72 and -53 contribute to the basal transcriptional activity of human LRP5 promoter. (A) Schematic representation of the KLF15 and Sp1 elements. Point mutations (underlined) were introduced to change the binding sites. (B) Constructs with native (pWT-72) or mutant KLF15 and/or Sp1 sites were transfected into HEK293 and U2OS cells, and the luciferase activity was determined. The luciferase activity of pWT-72 was arbitrarily set to 100%, and the activities of other constructs were calculated accordingly. (C and D) Sp1 and KLF15 transactivated the LRP5 promoter only if Sp1 and KLF15 binding sites were present. SL2 cells were cotransfected with either wild type construct (pWT-72) or mutated constructs (pMT1-72 or pMT2-72) along with the Sp1 (3C) or KLF15 (3D) expression constructs, and the relative luciferase activity was determined. The luciferase activity cotransfected with control vector (empty vector, EV) was set to 100%, and the relative activity under KLF15 or Sp1 stimulation was calculated accordingly. (E) Chromatin immunoprecipitation assay of KLF15 and Sp1 binding to human LRP5 promoter. The bindings of KLF15 and Sp1 to the human HSD17B5 promoter were used as a positive control (upper panel). The bindings of KLF15 to the LRP5 promoter (lower right panel) and the binding of Sp1 to the LRP5 promoter (lower left panel) were determined by ChIP using anti-KLF15 and anti-Sp1 antibodies, respectively. Anti-IgG antibodies were used as a negative control. The associated chromatin DNA fragments were amplified with the primer pairs flanking the Sp1 and KLF15 binding sites. Chromatin DNA input as described in the material and methods was subjected to the same PCR amplification.