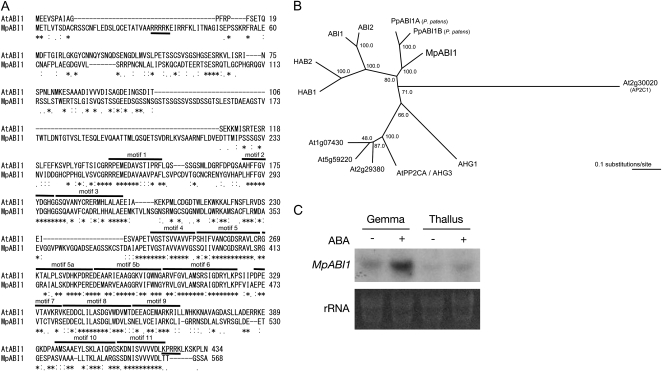
Figure 2.
Molecular characteristics of MpABI1. A, Sequence alignment of MpABI1 and Arabidopsis ABI1. The alignment was made by the T-COFFEE program. Identical amino acids are marked with asterisks, very similar ones are marked with colons, and weakly similar ones are marked with dots. Conserved motifs 1 to 11 of PP2Cs assigned by Bork et al. (1996) are indicated by overlines. Putative NLSs are underlined. B, Phylogenetic representations of MpABI1, PpABI1A, PpABI1B, and Arabidopsis ABI1-related PP2Cs. The phylogenetic tree was built on the alignment of amino acid sequences of the PP2C domains using the ClustalW program. The Arabidopsis Ap2C1 phosphatase sequence was used as the outgroup. The bar indicates 0.1 substitutions per site. C, Northern analysis of M. polymorpha mRNA for examination of MpABI1 expression. Total RNAs isolated from ABA-treated (+) or nontreated (−) gemma and thallus were electrophoresed, blotted onto a nylon membrane, and hybridized with a 32P-labeled MpABI1 cDNA probe. Ethidium bromide-stained rRNA of each sample is shown to verify equal loading.