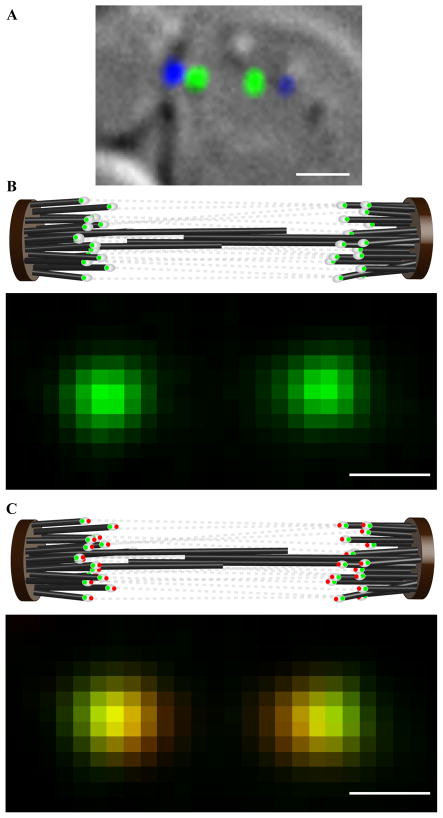
Figure 1. Measuring the distance separating two kinetochore proteins in a budding yeast metaphase spindle.
(a) A budding yeast cell in metaphase (DIC) with fluorescently labeled kinetochores (green) and spindle pole body (blue). (b) The cartoon depicts arrangement of kinetochores tagged with EGFP (white ovals with green dots) within the metaphase spindle. Tense chromatin connections (gray dotted lines) between sister kinetochores align them closely with the spindle axis. When such a cell is visualized with wide-field fluorescence microscopy, the two kinetochore clusters (each containing 16 kinetochores) appear as nearly diffraction-limited spots. (c) Metaphase spindle in a strain that has two kinetochore proteins, one protein fused to EGFP (green dots) and the other with tdTomato (red dots). When such cells are imaged simultaneously in the EGFP and tdTomato channels (lower panel), the offset between the centroids of the EGFP and tdTomato images of a kinetochore cluster can be used to determine the average distance separating the ends of the two proteins accurately. (scale bar ~ 1μm in a, ~ 500 nm in b and c; 1 pixel ~ 107 nm in b and c).