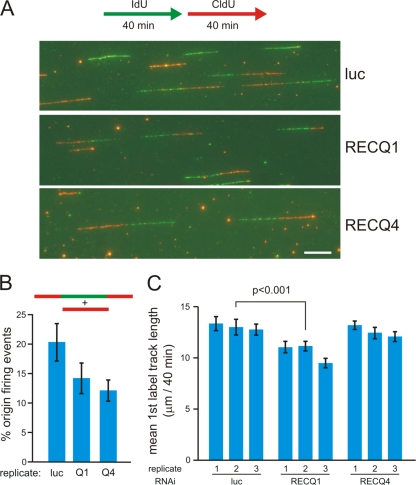
FIG. 9.
Depletion of human RECQ1 or RECQ4 affects DNA replication dynamics. (A) An outline of the experimental protocol is shown at the top, in which asynchronous cells were labeled consecutively with IdU (green) and then with CldU (red) for 40 min each prior to isolating and stretching DNA for immunostaining as described in Materials and Methods. Shown below this are representative images of replication tracks in control (luciferase)-, RECQ1-, and RECQ4-depleted cells. All track photos are shown at identical magnifications, where the white scale bar (lower right) is 10 μm long, representing approximately 39 kb of DNA. (B) RECQ1 or RECQ4 depletion reduces the probability of origin firing in stretched DNA samples. Origin firing events among all tracks labeled during the protocol for panel A were identified as CldU-only (red only) or CldU-IdU-CldU (red-green-red) triple-segment tracks (see diagram at top). The mean percentages of new origin firing events defined by these two track types among all labeled tracks are shown for three independent experiments in which 200 to 450 tracks/experiment were typed for control-, RECQ1-, or RECQ4-depleted cells. Error bars show standard deviations. The P values, calculated using Student's t test, are 0.074 between luciferase and RECQ1 and 0.021 between luciferase and RECQ4. (C) RECQ1 depletion, but not RECQ4 depletion, slows ongoing replication forks. The bar graph summarizes mean lengths of first-label IdU (green) segments labeled for 40 min in two-segment (green-red) tracks to ensure that fork rate measurements were made from active replication forks (see the diagram in panel A). Track lengths were measured in μm, using AxioVision software (Carl Zeiss), for three independent experiments in which 150 to 370 first-segment track lengths were measured for each sample. Error bars show 95% confidence intervals for sample means. The statistical significance of differences in mean track lengths was determined by a two-sample Kolmogorov-Smirnov test. A representative P value is shown for a control (luciferase)- versus RECQ1-depleted sample pair.