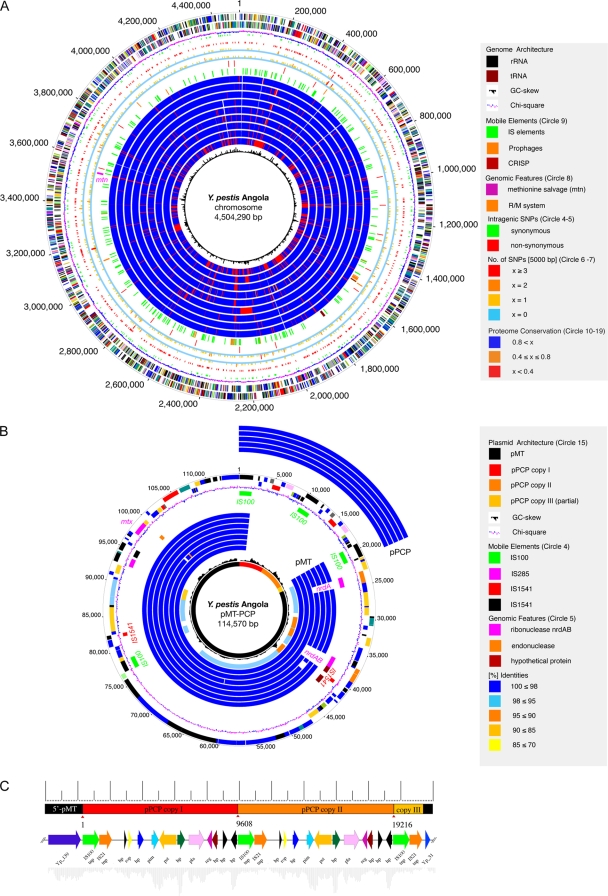
FIG. 1.
(A) Circular representation of the Y. pestis Angola chromosome. Circles, from outer to inner: predicted open reading frames encoded on the plus (1) and minus strands (2), colored according to the respective MANATEE roles; (3) G+C skew; (4) sSNP; (5) nsSNP; (6 and 7) genome-wide distribution of SNPs; (8) chromosomal regions of interest; (9) mobile elements; (10 to 18) comparative analysis of the Angola proteome to strains CO92 (10), KIM (11), Antiqua (12), Nepal516 (13), 91001 (14), and Pestoides F (15); interspecies comparison to Y. pseudotuberculosis strains IP32953 (16) and IP31758 (17) and Y. enterocolitica 8081 (18); (19) chi-square values. (B) Chimeric plasmid pMT-PCP. Circles, from outer to inner: (1 and 2) predicted open reading frames; (3) G+C skew; (4) mobile elements. Circle fragments for comparison to the pPCP plasmid are on the outside and show Y. pestis strains CO92 (1), KIM (2), 91001 (3), Antiqua (4), and Nepal516 (5); circle fragments for comparison to the pMT plasmids are on the inside and show strains CO92 (6), KIM (7), 91001 (8), Antiqua (9), Nepal516 (10), Pestoides F (11), and G8787 (12); circles for interspecies comparison to S. enterica CT18 pHCM2 plasmid (13); (14) chi-square values; (15) chimeric plasmid composition. Plasmid regions secondarily lost in the Y. pestis evolution, such as ribonucleases nrdAB and the Nepal516-specific 1,220-bp deletion of four hypothetical proteins with no assigned function (brown). (C) Architecture of pMT-PCP. The plasmid features a pPCP dimer integrated into the pMT plasmid in a head-to-tail arrangement.