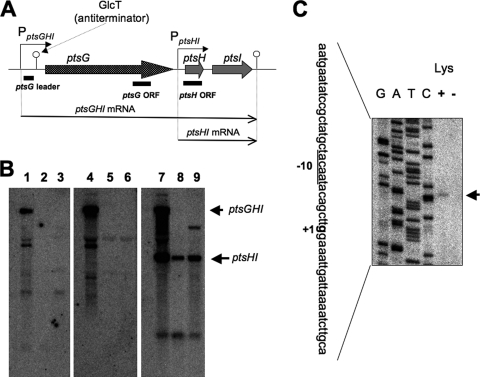
FIG. 2.
Downregulation of ptsGHI transcription upon lysine starvation and decoyinine treatment. (A) The ptsGHI operon has two promoters, PptsGHI for ptsGHI transcription and PptsHI for ptsHI transcription (45). GlcT, an antiterminator belonging to the BglG/SacY family, binds to a ribonucleic antiterminator upstream of ptsG in the presence of glucose. This prevents formation of an overlapping transcriptional terminator. The ptsGHI and ptsHI transcription terminator is also located downstream of ptsI. (B) Northern analysis of the stringent control of ptsGHI transcription. RNA samples were prepared from cells of strain 1A765 (lys) that had been subjected to lysine starvation for 30 min (lanes 2, 5, and 8) or to decoyinine treatment (500 μg/ml) for 30 min (lanes 3, 6, and 9) and from cells that were not subjected to these stresses (lanes 1, 4, and 7). The transcripts in the left, middle, and right panels were detected with the probes for the pstG leader, ptsG ORF, and ptsH ORF (indicated in panel A), respectively, whose preparation is described in Materials and Methods. The positions of the ptsGHI and ptsHI transcripts are indicated by the arrows on the right. (C) Primer extension analysis for mapping of the 5′ end of the ptsGHI transcript. Total RNA from cells of strain 1A765 (lys) which had been subjected to lysine starvation for 30 min (lane −) and total RNA from cells of strain 1A765 which had not been subjected to this nutrient stress (lane +) were annealed with the PptsG-R primer (see Table S1 in the supplemental material), and then primer extension was performed as described in Materials and Methods. Lanes G, A, T, and C contained the products of the corresponding dideoxy sequencing reactions with the PCR product as the template, as described in Materials and Methods. The part of the nucleotide sequence of the coding strand corresponding to the ladder is shown on the left; the transcription initiation base (+1) is indicated by bold type, and the corresponding −10 region for the ptsGHI promoter is underlined. The position of the runoff cDNA is indicated by the arrow on the right.