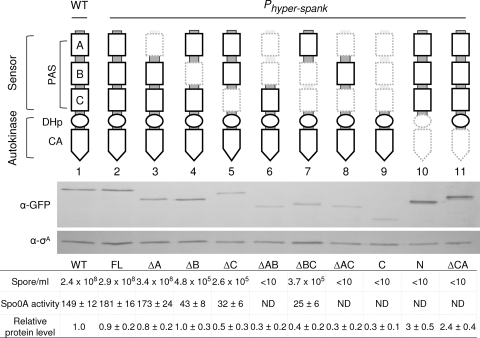
FIG. 1.
Quantitative assay for detection of the enzymatic activities of the KinA domain-deletion mutants. The schematic diagram at the top shows a domain organization of KinA. The sensor domain at the N terminus is further subdivided into three PAS domains (PAS-A, PAS-B, and PAS-C). The autokinase domain at the C terminus is further subdivided into a DHp and a CA domain (18, 25). The catalytic histidine (His405) is located within the DHp domain. Deleted portions are depicted in hatched lines in the diagrams. Genes for full-length KinA and each truncated mutant protein were fused to Phyper-spank and introduced at the amyE locus of cells containing both kinA and kinB null mutations. β-Galactosidase activities (Miller units) from PspoIIG-lacZ were measured at hour 3 after suspension as a measure of Spo0A activity. The same culture was used for the determination of sporulation efficiency. The wild-type strain (WT; lacking the IPTG-inducible construct) was used as a control. Strains used are as follows: MF3366 (WT), MF3316 (full-length KinA; FL), MF3317 (ΔPAS-A; ΔA), MF3318 (ΔPAS-B; ΔB), MF3319 (ΔPAS-C; ΔC), MF3320 (ΔPAS-AB; ΔAB), MF3321 (ΔPAS-BC; ΔBC), MF3322 (ΔPAS-AC; ΔAC), MF3323 (KinAC; C), MF3324 (KinAN; N), and MF3325 (ΔCA). Expression of full-length KinA or KinA mutant protein was examined by immunoblot analysis. Cell extracts of strains expressing GFP fusion proteins were prepared at hour 2 after suspension and were processed for immunoblotting with anti-GFP (α-GFP) antibodies. σA, a constitutively expressed protein, was used as a loading control. The protein levels were quantified and normalized to both the levels of σA and the levels of the wild-type KinA-GFP. Strains used for immunoblotting are as follows: MF3593 (WT; lane 1), MF3352 (FL; lane 2), MF3353 (ΔA; lane 3), MF3354 (ΔB; lane 4), MF3355 (ΔC; lane 5), MF3356 (ΔPAS-AB; lane 6), MF3357 (ΔPAS-BC; lane 7), MF3358 (ΔPAS-AC; lane 8), MF3359 (KinAC; lane 9), MF3360 (KinAN; lane 10), and MF3361 (ΔCA; lane 11). All experiments were performed at least three times independently. The representative data are shown. ND, not detected.