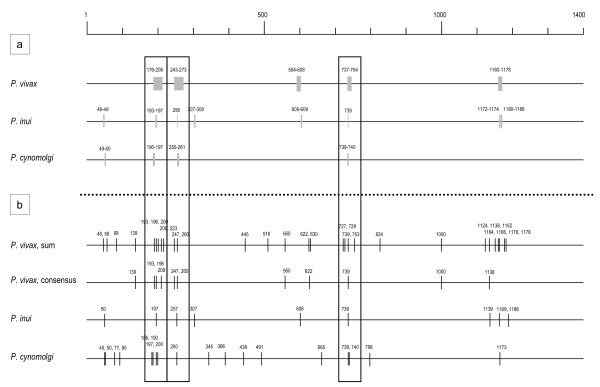
Figure 3.
Positively selected codon sites along the msp1 alignment of P. vivax and P. vivax-related simian malaria parasites. Positively selected sites with ω (= dN/dS) >1 were detected by (a) omegaMap [29] and (b) HyPhy/Datamonkey [30]. Boxes indicate positively selected regions shared by three parasite species. In (a), positions of codon showing ω > 1 with more than 95% posterior probability are denoted by half-tone bars. In (b), individual sites with ω significantly greater than 1 are mapped by vertical lines. In P. vivax, computations were repeated five times after random sampling from a pool of sequences, and sites detected at least once in the analyses; or those detected 3 times or more are shown as "sum" and "consensus", respectively. Refer to Additional file 3; figure S1 for individual amino acid replacements.