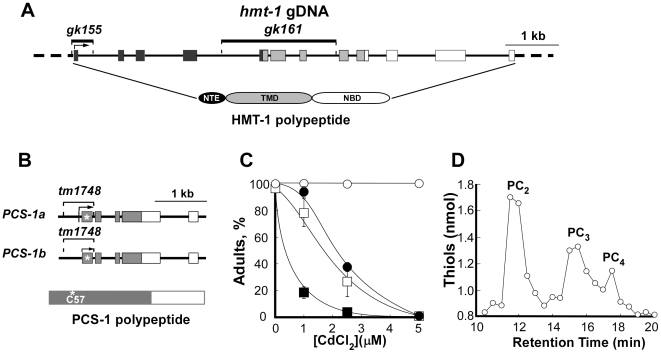
Figure 1. Molecular and functional analyses of hmt-1 and pcs-1 deletion alleles.
A. Structure of the hmt-1 gene, location of deletion mutations and domains of CeHMT-1 polypeptide. The black boxes depict regions encoding the N-terminal extension (NTE), light gray boxes depict regions encoding the transmembrane domain (TMD), and white boxes depict regions encoding the nucleotide-binding domain (NBD). The positions of gk155 and gk161 deletions are represented as lines above the genomic structure. B. Structure of the pcs-1 gene, location of the deletion mutation and domains of the PCS-1 polypeptide. The grey boxes represent regions that encode a conserved N-terminal part of PCS-1. An asterisk indicates a catalytic Cys57 residue of the conserved catalytic triad of the PCS-1 polypeptide. PCS-1a and PCS-1b are predicted splice variants of PCS-1. In A and B, the exons and the introns are depicted as boxes and as connecting lines, respectively. C. Cd hypersensitivity of hmt-1 and pcs-1 deletion mutants. Wild-type N2 (○), hmt-1(gk155) (•), hmt-1(gk161 (□) and pcs-1(tm1748) (▪) adult hermaphrodites were placed individually onto NGM plates with indicated concentrations of Cd and allowed to lay eggs. Shown are the percentages of the progeny that had reached adulthood 4 days after hatching. The total number of worms used for each strain and condition, and statistical significance of measurements are shown in Table S1. D. Reverse-phase HPLC analysis of phytochelatins in lysates prepared from Cd-grown N2 wild-type worms. The peaks designated “PC2”, “PC3” and “PC4” were identified on the basis of their co-migration with the in vitro synthesized PC standards [41].