Table 1.
Summary of candidate overlapping CDSs
| genus (RNA) |
phylogenetic distribution (GenBank RefSeqs) |
length (codons) |
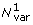 |
ln(LR)2 | 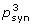 |
|---|---|---|---|---|---|
|
Mamastrovirus (sgRNA) |
Human, porcine and feline astroviruses etc [GenBank: NC_001943] |
91-122 | >500 | NA | 5.9 × 10-28 |
|
Seadornavirus (segment 7) |
Banna virus, Kadipiro virus etc [GenBank: NC_004204, GenBank: NC_004209] |
52-65 | 31 | 9.6 | 2.3 × 10-10 |
|
Cytorhabdovirus (P mRNA) |
Lettuce necrotic yellows virus etc [GenBank: NC_007642, GenBank: NC_011532] |
97-102 | 149 | 22.7 | NA |
|
Gammacoronavirus (NS6 sgRNA) |
Group 3c coronaviruses [GenBank: NC_011548, GenBank: NC_011549, GenBank: NC_011550] |
69-89 | 75* | 20.6* | NA |
|
Alphacoronavirus (ORF3 sgRNA) |
Bat coronaviruses 1A, 1B, HKU8 [GenBank: NC_010436, GenBank: NC_010437, GenBank: NC_010438] |
91-95 | 82* | 12.4* | NA |
1. Approximate total number of phylogenetically independent base variations in the ORFX region (not including sequences with only partial coverage of ORFX).
2. Total MLOGD 'log likelihood' score (not strictly log likelihood due to non-independence issues - see Ref. [1] for details). Positive values indicate that ORFX is likely to be coding. Candidates that were not detected with MLOGD are indicated with 'NA'. For candidates marked with an '*', the poorly constrained C-terminal region (see Figures 7, 9) was excluded for calculation of Nvar and ln(LR).
3. Probability that the degree of conservation at synonymous sites of the known CDS within the whole ORFX region could be obtained under a null model of neutral evolution at synonymous sites. Note that a significant p-value here does not necessarily indicate an overlapping CDS, merely an overlapping functional element. Candidates for which the available sequence data are too limited to effectively use this approach are indicated with 'NA'.
