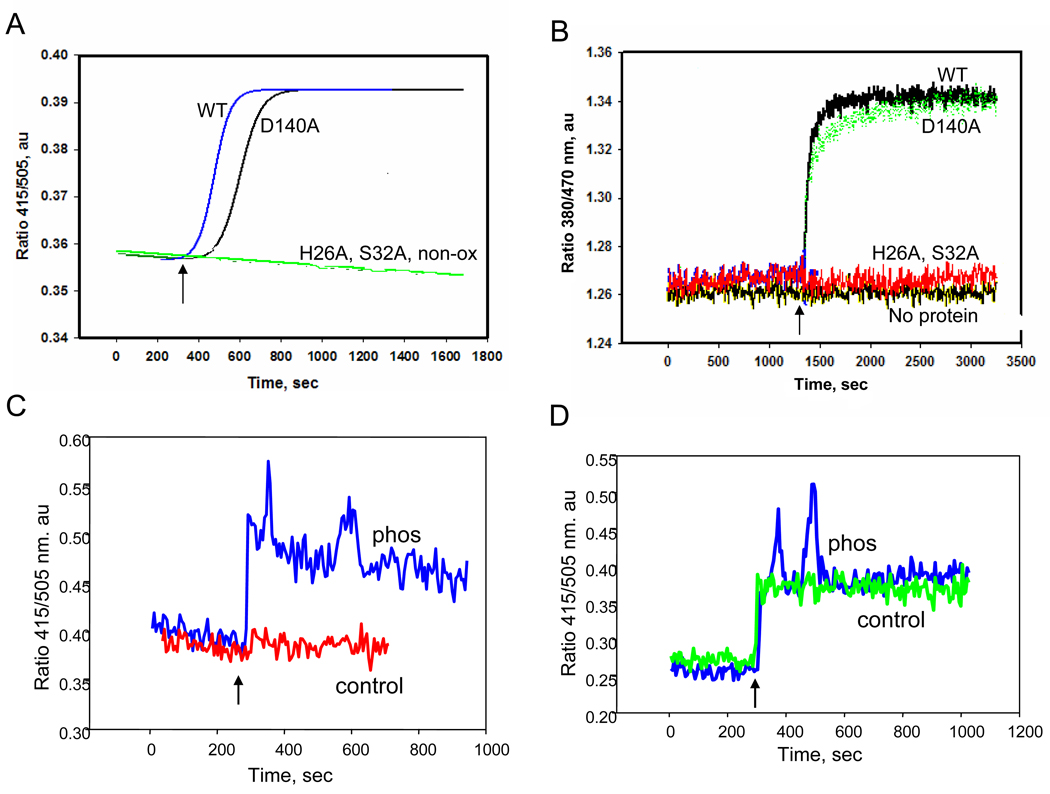
Figure 5. Fluorescence analysis of Prdx6 to binding liposomes.
Prdx6, phosphorylated Prdx6, and Prdx6 mutants were evaluated for binding to liposomes containing all reduced phospholipids or liposomes that were oxidized by treatment with Cu2+/ascorbate. Prdx6 (3.2 µM) was incubated in 40 mM PBS at pH 7.4 and liposomes (100µM) were added. A. Kinetics of Prdx6 binding to unilamellar liposomes labeled with 2 mol % DNS-PE. The ratio of fluorescence emission at 415/505 nm is shown as a function of time following the addition of lipid (indicated by the arrow). The binding kinetics are shown for addition of oxidized liposomes to wild type (WT) Prdx6 or in the presence of the Prdx6 mutants S32A, D140A or H26A. The binding curve for wild type Prdx6 with non-oxidized liposomes (non-ox) overlapped the H26A and S32A curves. B. Incubation as in A but with bis-Pyr-PC (2 mol%) in oxidized unilamellar liposomes. Fluorescence emission was recorded as the ratio of 380/470 nm for wild type (WT) Prdx6 and S26A, H26A and D140A mutants. ‘No protein’ indicates a control tracing in the absence of Prdx6. C. Fluorescence emission ratio for DNS-PE labeled liposomes (non-oxidized) in the presence of phosphorylated (Phos) or non-phosphorylated (control) wild type Prdx6. D. Same experiment as C but with oxidized liposomes.