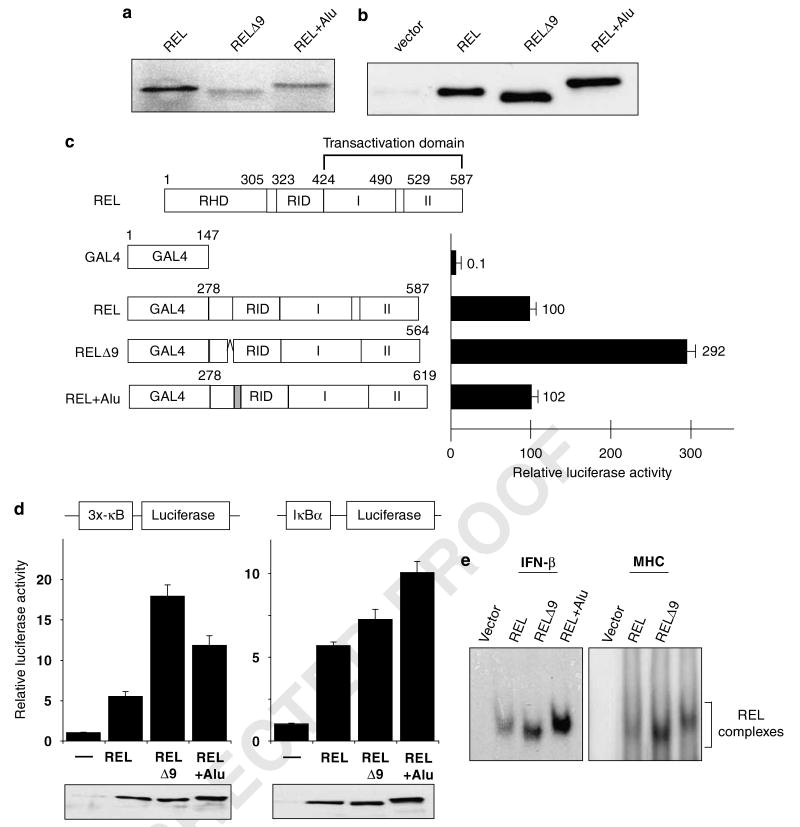
Figure 6.
REL isoforms differentially affect REL-mediated transcriptional activation and κB site DNA binding. (a) pcDNA3.1 plasmids containing the indicated REL sequences were subjected to in vitro translation in the presence of [35S]methionine. Proteins were separated by SDS-polyacrylamide gel and detected by phosphorimaging. (b) A293 cells were transiently transfected with pcDNA 3.1 expression vectors containing the indicated REL cDNAs. The cell extracts were subjected to anti-REL western blotting. (c) Shown at top is a schematic of full-length REL. GAL4 fusion proteins containing the indicated REL C-terminal sequences were analysed for their ability to activate transcription from a GAL4 site-containing luciferase reporter locus in A293 cells, as described for Figure 1. Values are relative to that seen with GAL4–REL (100). (d) Reporter gene assays with the indicated κB site-containing luciferase cassettes were performed in A293 cells as described for Figure 3a. Luciferase activities are relative to pcDNA (1.0), and values are the averages of three independent experiments performed in triplicate. In the bottom panels, extracts from triplicate samples used in the reporter assays were analysed by anti-REL western blotting. (e) An electrophoretic mobility shift assay was performed with whole-cell extracts from A293 cells transiently transfected with expression vectors containing each of the three REL splice variants. The κB site-containing probe used is shown at the top. The REL complexes are indicated. RHD, Rel homology domain; RID, REL inhibitory domain; I and II, transactivation subdomains I and II.