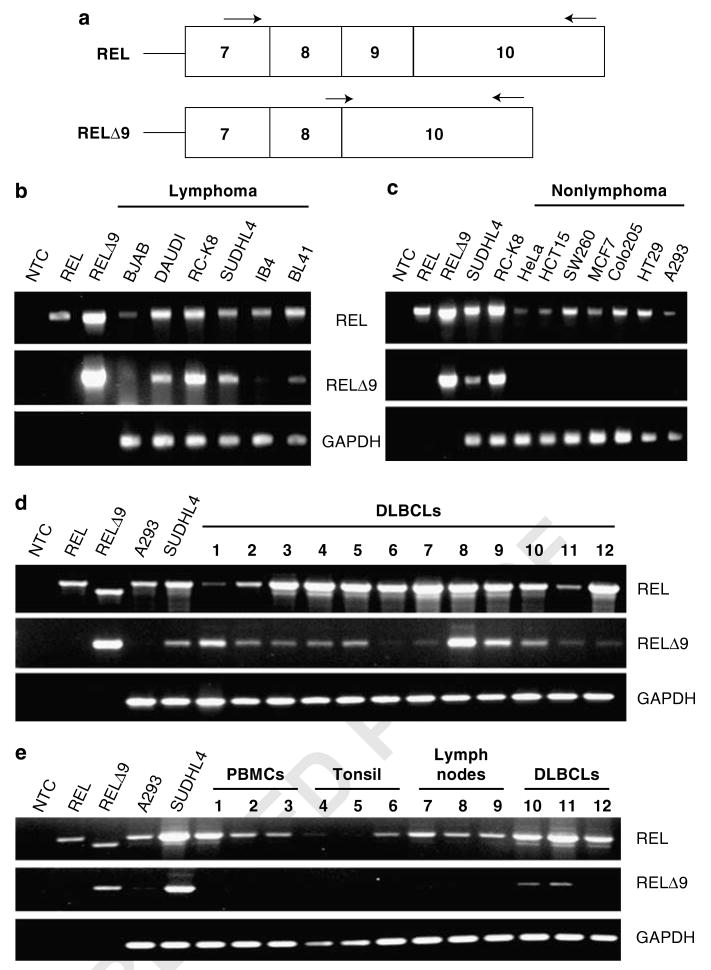
Figure 7.
The RELΔ9 isoform shows increased expression in lymphoma cell lines. (a) General structure of the relevant section of the REL cDNAs. Arrows indicate approximate positions of RT–PCR primers used. (b) RT–PCR detection of mRNAs for the REL and RELΔ9 transcripts in a panel of lymphoma cell lines. (c) RT–PCR detection of the REL and RELΔ9 transcripts in a panel of nonlymphoma human cell lines: HeLa, cervical cancer; HCT15, colon carcinoma; SW260, colon carcinoma; MCF7, breast cancer; Colo205, colorectal cancer; HT29, colon adenocarcinoma; and A293, adenovirus-transformed embryonic kidney. (d) RT–PCR detection of the REL and RELΔ9 transcripts in a panel of primary human diffuse large B-cell lymphoma (DLBCL) samples. (e) Comparison of RT–PCR of the REL and RELΔ9 transcripts from DLBCLs (samples 8, 9 and 12, as in d) with primary human, non-neoplastic samples from peripheral mononuclear blood cells (PMBCs), tonsil and lymph nodes. The REL-specific forward primer is in exon 7, the RELΔ9-specific forward primer spans the exon 8/exon 10 boundary and the reverse primer for amplification of both REL and RELΔ9 anneals within exon 10. PCRs of SUDHL4 and RC-K8 cDNAs are used as positive controls for amplification of the RELΔ9 splice variant cDNA. NTC, no template control. PCRs of pcDNA–REL and pcDNA–RELΔRID plasmids were used as controls for product size and primer specificity. GAPDH is included as a control for cDNA quantity.