Fig. 19.
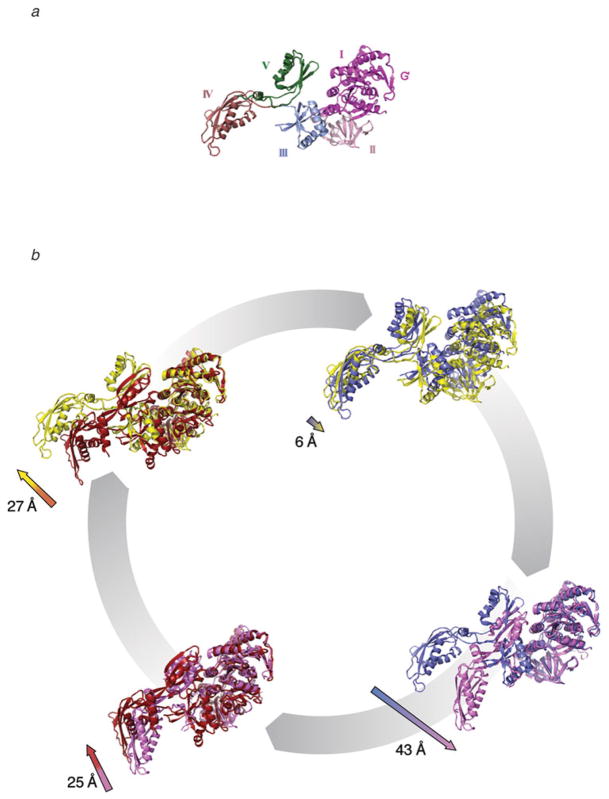
Comparison of structural changes observed in EF-G. (a) The conformational changes observed in EF-G involve a hinge-like movement of the C-terminal domains (III, IV, and V) with respect to the N-terminal domains (I, II, and G′). (b) The conformations observed in EF-G/eEF2 by means of cryo-EM (ribosome-bound forms) and X-ray crystallography (in solution) can be summarized as follows: EF-G in complex with GTP in solution (red), before interacting with the ribosome, as seen in the crystal structure of the EF-G analog EF-G-2 (PDB code: 1WDT). When EF-G•GTP binds to the ribosome, the conformational change of EF-G causes a shift in the tip of domain IV (structure show in yellow). EF-G in the new conformation likely stabilizes the ribosome in the ratcheted conformation. This EF-G•GTP•ribosome conformation was determined by cryo-EM using the nonhydrolyzable GTP analog GDPNP (PDB code: 1PN6). The conformational changes in ribosome-bound EF-G, upon GTP hydrolysis, are likely similar to those described for eEF2•80S complexes in Taylor et al. (2007). A comparison of the complexes before (yellow) and after GTP hydrolysis (blue, PDB code: 2P8Y) reveals small magnitude shifts in domains I, II, and G′ toward the GAC of the ribosome, a reorganization of the Switch I loop, and an ~6-Å shift in domain IV toward the decoding center of the ribosome. This movement is thought to sever the connection between the decoding center in the body of the small subunit and the A-site mRNA–tRNA duplex bound to the head of the small subunit, so that movement of the mRNA–tRNA complex can occur via a head rotation of the small subunit. Conformational changes of EF-G•GDP from the ribosome-bound conformation (blue), determined by cryo-EM (PDB code: 2P8Y), to the solution structure (pink), determined by X-ray crystallography (PDB code: 1EFG), include a movement at the tip of domain IV by ~43 Å. This conformational change in EF-G likely contributes to its dissociation from the ribosome. A comparison of the GDP-bound conformation (pink) of EF-G with the GTP-bound conformation (red) reveals a shift at the tip of domain IV of the factor by ~25 Å. The presence of GTP also reveals an ordered Switch I loop and a different conformation of the Switch II loop. Altogether, these rearrangements likely are the basis of a much higher affinity of the pre-translocational ribosome for the GTP-bound conformation of EF-G. This figure was altered for the purposes of this article. Data adapted from Frank et al. (2007), copyright (2007) National Academy of Sciences.