Fig. 20.
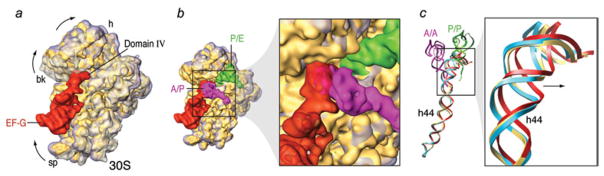
Kinetic efficiency of the hybrid configuration. (a) Superimposition of aligned 30S subunits from hybrid (solid yellow), classic (transparent gray), and EF-G-bound (transparent purple) ribosomes. The binding position of EF-G (red) is shown for illustration purposes. (b) Close-up of the superimposition of hybrid (solid yellow) and EF-G-bound (transparent purple) ribosomes in the presence of EF-G•GDPNP and hybrid tRNAs. (c) Superimposed atomic models of 16S rRNA h44 for the classic state ribosome (blue), the hybrid state ribosome (yellow) and the EF-GGDPNP bound ribosome (red). The lower portion of h44 has been kept fixed in the fitting to show the displacement of the decoding region (top). The arrow points in the direction of the movement. A/A and P/P tRNAs are shown for clarity. This figure was altered for the purposes of this article. Data adapted from Agirrezabala et al. (2008), copyright (2008) Cell Press.