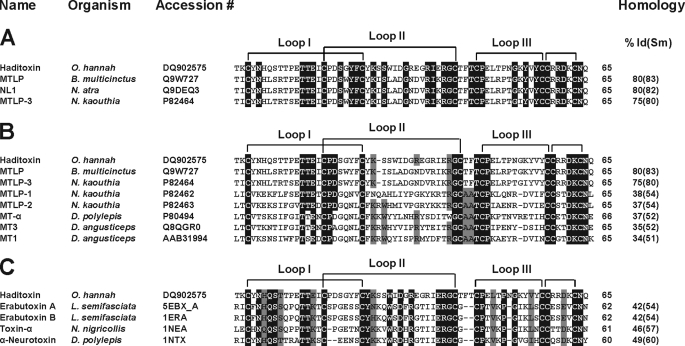
FIGURE 1.
Multiple sequence alignment of novel proteins. A–C, sequence alignment of haditoxin with the most homologous sequences (A), muscarinic toxin homologs (B), and short-chain α-neurotoxins (C). Toxin names, species, and accession numbers are shown. Conserved residues in all sequences are highlighted in black. Disulfide bridges and loop regions are also shown. At the end of each sequence, the numbers of amino acids are stated. The homology (sequence identity and similarity (% Id(Sm))) of each protein is compared with haditoxin and shown at the end of each sequence. N. atra, Naja atra; N. kaouthia, Naja kaouthia; D. polylepis, Dendroaspis polylepis; and L. semifasciata, Laticauda semifasciata.