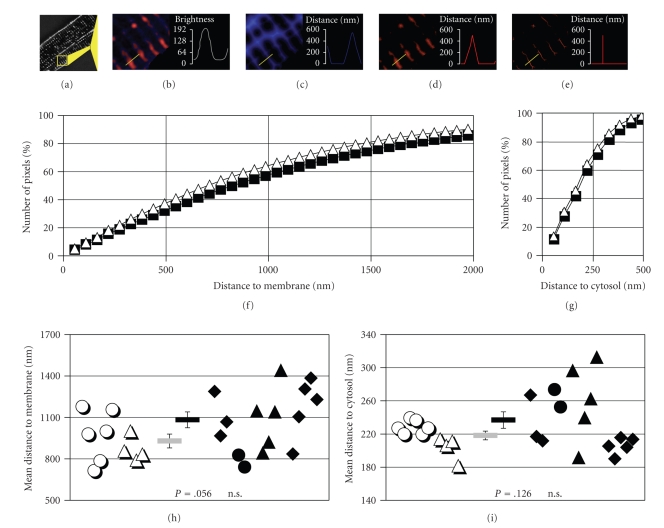
Figure 5.
Distance Distribution of the Transverse-Axial Tubule System. Every image of a cell ((a), scaled up and colored in (b)) was transferred into Euclidian distance maps. The distance from any pixel representing cytosol (blue) to its next neighboring membrane (red) within the image was analyzed and then plotted as a brightness value in the distance map (c). The Euclidian distance maps, of the TATS were developed within 3D recontructions of the TATS (d) and reduced to their 3D-reconstructed skeleton in order to analyze regional object diameters (e). From cytosolic distance maps the average of 22 × 106 pixels (non-failing) versus 50 × 106 pixels (failing) was plotted as normalized number of pixels against distance to membrane (f). From TATS distance maps, the average of 2.1 × 106 pixels (non-failing) versus 4.4 × 106 pixels (failing) was plotted as normalized number of pixels against distance to the next neighboring cytosolic pixel (g). Weighted means of the measurements resolve the mean distance between cytosol and membrane (h), and respectively, the mean object diameter (i). Open symbols represent measurements from non-failing hearts (2 hearts, 10 cells, 467 slices), closed symbols from failing hearts (4 hearts, 15 cells, 872 slices). Different symbols denote different cell batches. Bar symbols in the middle of chart (h) and (i) indicate the mean values ± SEM. Calculated P-values as given. Exemplary distance maps (b)–(d) are shown brightened up for improved illustration.