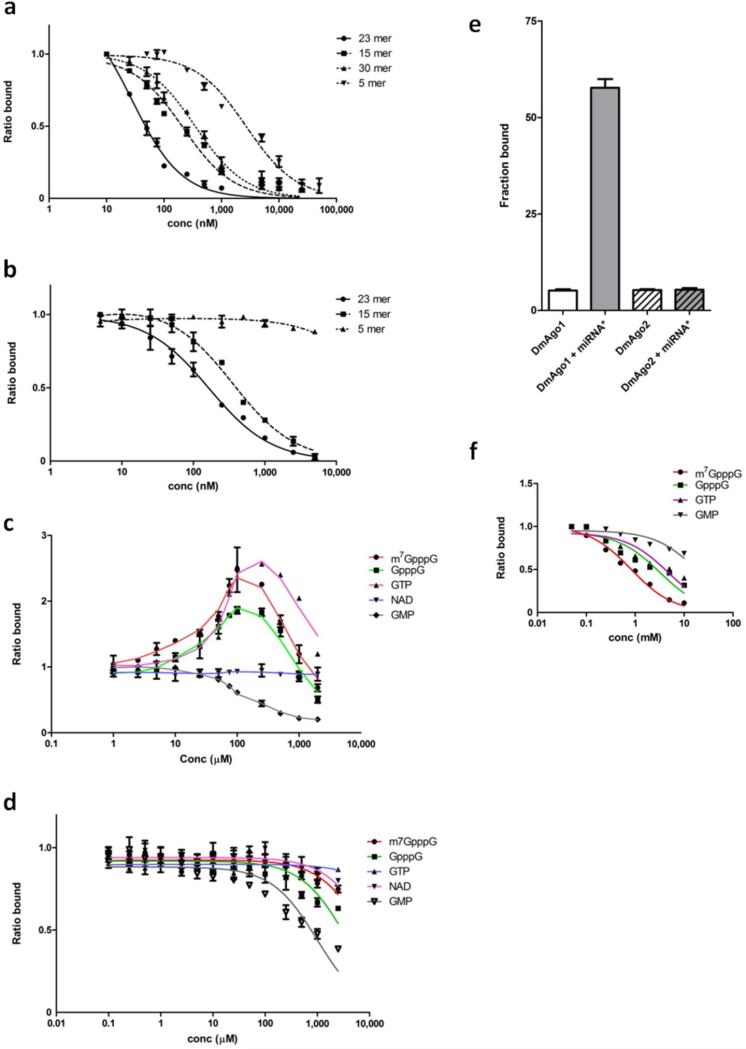
Figure 3.
Filter binding assays indicate two allosterically regulated nucleotide binding sites. Binding of 32P-labeled bantam to DmAgo1 in a and DmAgo2ΔQ in b in the presence of non-labeled RNAs of varying lengths (5, 15, 23 and 30 nts). Competition between labeled and non-labeled RNAs was performed by titration of the non-labeled species and represented by the ratio of bound 32P-bantam in the presence and absence of competing RNA species. Influence of different nucleotide substrates on the 32P-bantam binding to DmAgo1 in c and DmAgo2ΔQ in d. Nucleotide substrates were titrated and the amount of the bound 32P-bantam was followed. Values represent the ratio of bound 32P-bantam in the presence and absence of indicated nucleotide substrate. e, Binding of bantam duplex (miRNA*) to DmAgo1 and DmAgo2ΔQ stimulates only binding of DmAgo1 to 3H-labeled m7GpppG mRNA. f, Influence of different nucleotide substrates on binding of 3H-m7GpppG mRNA to DmAgo1. Error bars represent the standard error from at least three experiments.