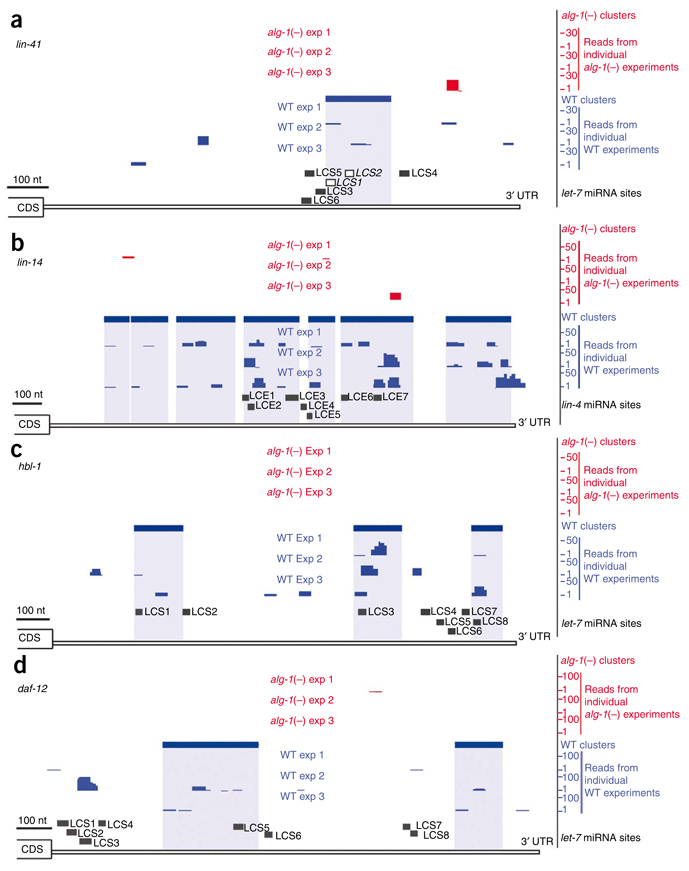
Figure 1.
MicroRNA targets identified by ALG-1 CLIP-seq in L4-stage worms. Graphical depictions of the number and location of reads from alg-1(−) (red upper tracks) and WT (blue lower tracks) from three biological replicates, CLIP-derived clusters (solid rectangular boxes over the reads) and putative miRNA binding sites in the 3′ UTR of mRNA transcripts (LCS, let-7 complementary sequence; LCE, lin-4 complementary element). (a) lin-41 3′ UTR. Of the six predicted LCSs, LCS1 and LCS2 (open boxes) have experimentally validated let-7 sites26–28. (b) lin-14 3′ UTR. Deletion of all the predicted LCE sites or of LCEs 1–5 results in misregulation of lin-14 expression23,29. (c) hbl-1 3′ UTR16,24,31. The sites for let-7 miRNA (LCSs 1–8) binding have been predicted but not experimentally tested. (d) daf-12 3′ UTR. Deletion of the predicted LCSs 1–4 or 5–8 in reporter constructs leads to misregulation of reporter gene expression18.