Figure 1.
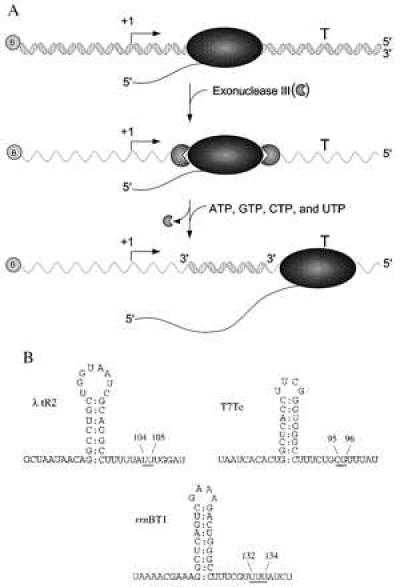
Diagram of experimental design and intrinsic terminator RNA secondary structures. (A) Transcription was initiated at the promoter (bent rightward arrow) and the ternary complexes were walked proximal to the termination site (T) on double-stranded PCR-generated templates that were biotinylated (shaded circle containing the letter B) at one of the 5′ ends. Next, the stalled elongation complexes (shaded ellipse) were digested with Exo III (shaded circles, missing a piece) and then all four NTPs were added, allowing the stalled elongation complex to resume transcript elongation. 5′ and 3′ ends of the DNA are as indicated. (B) The RNA secondary structures of the three intrinsic terminators, λ tR2, T7Te, and rrnBT1, are shown. The underlined bases indicate the positions of transcript release and the length of the RNA product [λ tR2 and rrnBT1 (ref. 24, T. Kerppola and M.J.C., unpublished results) and T7Te (25)]. The RNA secondary structures are drawn to conform to the classical model of transcript termination, with a G+C-rich stem and U-rich region immediately downstream. It should be noted, however, that for the λ tR2 and the rrnBT1 terminators, the hairpin can be extended to include upstream adenosines and downstream uridines.
