Table 2. Results of fitting evolutionary models for differences in 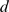 N/
N/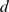 S ratios.
S ratios.
ER -LBD -LBD |
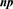
|

|
AIC | |
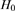 : Everyone is equal : Everyone is equal |
59 | −5645.4 | 11409 | |
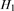 : O : O
 others others |
60 | −5641.7 | 11403 |

|
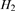 : O : O
 O O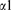
 others others |
61 | −5641.6 | 11405 | |
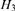 : O : O
 S S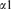
 others others |
61 | −5641.5 | 11405 | |
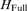 : Everyone is different : Everyone is different |
115 | −5610.3 | 11451 |
LBD and DBD indicate ligand and DNA binding domains, respectively. O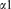 , O
, O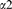 , O
, O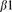 and O
and O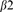 are the O. mykiss ER
are the O. mykiss ER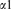 , ER
, ER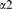 , ER
, ER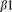 and ER
and ER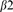 genes, respectively. S
genes, respectively. S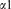 indicates all of the salmonid ER
indicates all of the salmonid ER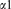 genes.
genes. 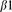 and
and 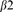 indicate ER
indicate ER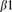 and ER
and ER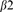 from all fish, respectively.
from all fish, respectively. 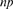 is the number of parameters in the model,
is the number of parameters in the model,  is the log likelihood calculated by PAML, and AIC is the Akaike Information Criterion value [35]. Models labeled with an asterisk are the best fitting models based upon the AIC values.
is the log likelihood calculated by PAML, and AIC is the Akaike Information Criterion value [35]. Models labeled with an asterisk are the best fitting models based upon the AIC values.
